Introduction
The calcofi4r package provides access to the CalCOFI
(California Cooperative Oceanic Fisheries Investigations) database,
which contains over 75 years of oceanographic and biological data from
the California Current ecosystem.
The database is organized into a tidy relational structure stored as Parquet files on Google Cloud Storage. This allows fast queries without downloading the entire database.
Connect to the Database
library(calcofi4r)
library(dplyr)
library(DBI)
library(sf)
library(mapview)
# connect to the latest CalCOFI database release
con <- cc_get_db()
# list available tables
dbListTables(con)
#> [1] "bottle" "bottle_measurement" "cast_condition"
#> [4] "casts" "cruise" "grid"
#> [7] "ichthyo" "lookup" "measurement_type"
#> [10] "net" "segment" "ship"
#> [13] "site" "species" "taxa_rank"
#> [16] "taxon" "tow"Convenience Functions
The package provides convenience functions for common operations:
# list available versions
cc_list_versions()
#> # A tibble: 1 × 6
#> version release_date tables total_rows size_mb is_latest
#> <chr> <chr> <int> <int> <dbl> <lgl>
#> 1 v2026.02 2026-02-05 17 13410422 80.9 TRUE
# list tables
cc_list_tables()
#> [1] "bottle" "bottle_measurement" "cast_condition"
#> [4] "casts" "cruise" "grid"
#> [7] "ichthyo" "lookup" "measurement_type"
#> [10] "net" "segment" "ship"
#> [13] "site" "species" "taxa_rank"
#> [16] "taxon" "tow"
# describe a table
cc_describe_table("ichthyo")
#> # A tibble: 7 × 3
#> column_name data_type is_nullable
#> <chr> <chr> <chr>
#> 1 species_id INTEGER YES
#> 2 life_stage VARCHAR YES
#> 3 measurement_type VARCHAR YES
#> 4 measurement_value DOUBLE YES
#> 5 tally INTEGER YES
#> 6 net_id INTEGER YES
#> 7 ichthyo_id INTEGER YES
# list measurement types
cc_list_measurement_types() |> head(10)
#> # A tibble: 10 × 3
#> measurement_type description units
#> <chr> <chr> <chr>
#> 1 alkalinity_rep1 Total alkalinity replicate 1 umol/kg
#> 2 alkalinity_rep2 Total alkalinity replicate 2 umol/kg
#> 3 ammonia Ammonia concentration (QC'd) umol/L
#> 4 barometric_pressure Barometric pressure millibars
#> 5 c14_dark 14C assimilation dark/control bottle mgC/m3/hld
#> 6 c14_mean Mean 14C assimilation of replicates 1 and 2 mgC/m3/hld
#> 7 c14_rep1 14C assimilation replicate 1 mgC/m3/hld
#> 8 c14_rep2 14C assimilation replicate 2 mgC/m3/hld
#> 9 chlorophyll_a Chlorophyll-a measured fluorometrically ug/L
#> 10 cloud_amount Cloud amount in oktas (WMO 2700) oktasRead Data Directly
Convenience functions return tibbles with optional filtering:
# read species data
species <- cc_read_species()
head(species)
#> # A tibble: 6 × 6
#> species_id scientific_name itis_id worms_id common_name gbif_id
#> <int> <chr> <int> <int> <chr> <int>
#> 1 1 Teleostei 161105 293496 Unidentified Teliost NA
#> 2 3 Elopidae 161109 153689 Tenpounders NA
#> 3 4 Elops affinis 161112 275403 Machete NA
#> 4 5 Albulidae 161119 151805 Bonefishes NA
#> 5 7 Albula 161120 157878 NA NA
#> 6 9 Clupeiformes 161694 10297 NA NA
# read ichthyo data (first 100 rows for demo)
ichthyo_sample <- cc_read_ichthyo() |> head(100)
head(ichthyo_sample)
#> # A tibble: 6 × 7
#> species_id life_stage measurement_type measurement_value tally net_id
#> <int> <chr> <chr> <dbl> <int> <int>
#> 1 1 egg NA NA 52 10
#> 2 206 larva NA NA 3 10
#> 3 288 larva NA NA 2 10
#> 4 301 larva NA NA 1 10
#> 5 31 larva NA NA 41 10
#> 6 31 larva size 10.8 1 10
#> # ℹ 1 more variable: ichthyo_id <int>
# read with filtering (lazy query)
anchovy <- cc_read_ichthyo(species_id == 19, collect = FALSE)
anchovy |> head(5) |> collect()
#> # A tibble: 5 × 7
#> species_id life_stage measurement_type measurement_value tally net_id
#> <int> <chr> <chr> <dbl> <int> <int>
#> 1 19 larva NA NA 2 1001
#> 2 19 larva size 11.8 1 1001
#> 3 19 larva size 8.8 1 1001
#> 4 19 egg NA NA 1 10017
#> 5 19 egg stage 11 1 10017
#> # ℹ 1 more variable: ichthyo_id <int>Database Schema
The CalCOFI database contains data from two main programs:
Ichthyoplankton Survey (since 1949):
-
cruise- cruise metadata (691 cruises) -
ship- research vessels (48 ships) -
site- sampling locations (61K sites) -
tow- net tow events (76K tows) -
net- net samples (77K nets) -
ichthyo- fish larvae counts (831K records) -
species- species taxonomy (1,144 species) -
taxon- taxonomic hierarchy -
grid- CalCOFI station grid (218 cells) -
segment- line segments between stations
Bottle Database (since 1949):
-
casts- CTD/bottle cast events (36K casts) -
bottle- water sample bottles (895K bottles) -
bottle_measurement- oceanographic measurements (11M records) -
measurement_type- measurement definitions (47 types) -
cast_condition- cast quality flags
# show row counts for each table
tables <- dbListTables(con)
tibble(
table = tables,
rows = sapply(tables, function(t) {
dbGetQuery(con, sprintf("SELECT COUNT(*) as n FROM %s", t))$n
})) |>
arrange(desc(rows))
#> # A tibble: 17 × 2
#> table rows
#> <chr> <dbl>
#> 1 bottle_measurement 11135600
#> 2 bottle 895371
#> 3 ichthyo 830873
#> 4 cast_condition 235513
#> 5 net 76512
#> 6 tow 75506
#> 7 site 61104
#> 8 segment 60413
#> 9 casts 35644
#> 10 taxon 1671
#> 11 species 1144
#> 12 cruise 691
#> 13 grid 218
#> 14 ship 48
#> 15 measurement_type 47
#> 16 taxa_rank 41
#> 17 lookup 26Query Bottle Data
The bottle data contains oceanographic measurements from CTD casts.
Measurements are stored in a normalized “long” format in
bottle_measurement.
# get temperature measurements with location and depth
d_temp <- dbGetQuery(con, "
SELECT
c.lon_dec as lon,
c.lat_dec as lat,
c.datetime_utc,
b.depth_m,
bm.measurement_value as temperature
FROM bottle_measurement bm
JOIN bottle b ON bm.bottle_id = b.bottle_id
JOIN casts c ON b.cast_id = c.cast_id
WHERE bm.measurement_type = 'temperature'
AND bm.measurement_value IS NOT NULL
AND b.depth_m <= 10
LIMIT 100000")
head(d_temp)
#> lon lat datetime_utc depth_m temperature
#> 1 -124.3500 37.28333 1950-06-14 18:48:00 0 12.98
#> 2 -124.3500 37.28333 1950-06-14 18:48:00 10 12.98
#> 3 -123.6167 37.61667 1950-06-15 01:18:00 0 13.59
#> 4 -123.6167 37.61667 1950-06-15 01:18:00 9 13.57
#> 5 -123.6167 37.61667 1950-06-15 01:18:00 10 13.56
#> 6 -123.1250 37.61667 1950-06-15 06:48:00 0 12.81
nrow(d_temp)
#> [1] 93985Summarize by Location
# summarize surface temperature by location
d_t <- d_temp |>
group_by(lon, lat) |>
summarize(
n = n(),
t_avg = mean(temperature, na.rm = TRUE),
.groups = "drop") |>
filter(!is.na(lon), !is.na(lat)) |>
st_as_sf(coords = c("lon", "lat"), crs = 4326, remove = FALSE)
head(d_t)
#> Simple feature collection with 6 features and 4 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -164.0833 ymin: 20.05 xmax: -150.025 ymax: 42.38333
#> Geodetic CRS: WGS 84
#> # A tibble: 6 × 5
#> lon lat n t_avg geometry
#> <dbl> <dbl> <int> <dbl> <POINT [°]>
#> 1 -164. 42 2 10.1 (-164.0833 42)
#> 2 -158. 42.4 16 16.7 (-157.9833 42.36667)
#> 3 -158. 42.4 4 11.8 (-157.9833 42.38333)
#> 4 -153. 20.1 4 24.2 (-153.1 20.10833)
#> 5 -150. 31.2 3 23.1 (-150.1167 31.2)
#> 6 -150. 20.0 4 24.3 (-150.025 20.05)CalCOFI Grid
The CalCOFI sampling grid defines standard station positions. The package includes pre-loaded grid data:
-
cc_grid- station polygons -
cc_grid_ctrs- station centroids -
cc_grid_zones- aggregated zones by station pattern
# show the CalCOFI grid colored by zone
mapview(cc_grid, zcol = "zone_key", layer.name = "Zone") +
mapview(cc_grid_ctrs, cex = 1, col.regions = "black", legend = FALSE)Grid from Database
The grid is also available in the database with additional attributes:
# query grid from database (includes geometry)
grid_db <- dbGetQuery(con, "SELECT * EXCLUDE(geom, geom_ctr) FROM grid")
head(grid_db)
#> grid_key station line shore pattern spacing
#> 1 st0-ln10_hist 0 10 nearshore historical 20
#> 2 st20-ln10_hist 20 10 nearshore historical 20
#> 3 st40-ln10_hist 40 10 nearshore historical 20
#> 4 st60-ln10_hist 60 10 nearshore historical 20
#> 5 st80-ln10_hist 80 10 offshore historical 20
#> 6 st100-ln10_hist 100 10 offshore historical 20
#> zone area_km2
#> 1 nearshore-historical 23111.47
#> 2 nearshore-historical 32060.98
#> 3 nearshore-historical 32467.41
#> 4 nearshore-historical 32869.45
#> 5 offshore-historical 33267.04
#> 6 offshore-historical 33660.13Show Effort by Grid Cell
Join temperature observations to the CalCOFI grid to show sampling effort:
# count observations per grid cell
n_grid <- cc_grid |>
st_join(d_t) |>
group_by(sta_key) |>
summarize(n = sum(n, na.rm = TRUE))
mapview(n_grid, zcol = "n", layer.name = "Observations")Show Effort by Station Point
# join counts to centroids
n_pts <- cc_grid_ctrs |>
left_join(
n_grid |> st_drop_geometry() |> select(sta_key, n),
by = "sta_key")
mapview(n_pts, cex = "n", layer.name = "Observations")Map Contours
Interpolate temperature data to create contour maps using Inverse Distance Weighting (IDW).
All Zones
# interpolate points to raster using IDW
r_all <- pts_to_rast_idw(d_t, "t_avg", cc_grid_zones)
# generate contour polygons
p_all <- rast_to_contours(r_all, cc_grid_zones)
mapview(p_all, zcol = "z_avg", layer.name = "Temp (C)")Standard and Extended Pattern
# filter to standard + extended zones
aoi_ext <- cc_grid_zones |>
filter(sta_pattern %in% c("standard", "extended"))
# interpolate and contour
r_ext <- pts_to_rast_idw(d_t, "t_avg", aoi_ext)
p_ext <- rast_to_contours(r_ext, aoi_ext)
mapview(p_ext, zcol = "z_avg", layer.name = "Temp (C)")Query Ichthyoplankton Data
The ichthyoplankton survey counts fish larvae by species across sampling sites.
# get top 10 species by total count
top_species <- dbGetQuery(con, "
SELECT
s.scientific_name,
s.common_name,
SUM(i.tally) as total_count,
COUNT(DISTINCT n.tow_id) as n_tows
FROM ichthyo i
JOIN species s ON i.species_id = s.species_id
JOIN net n ON i.net_id = n.net_id
GROUP BY s.scientific_name, s.common_name
ORDER BY total_count DESC
LIMIT 10")
top_species
#> scientific_name common_name total_count n_tows
#> 1 Teleostei Unidentified Teliost 8960182 61416
#> 2 Engraulis mordax Northern anchovy 8553961 29491
#> 3 Sardinops sagax Pacific sardine (pilchard) 1430924 9766
#> 4 Merluccius productus Pacific hake or whiting 1298807 12527
#> 5 Vinciguerria lucetia Panama lightfish 500036 14829
#> 6 Sebastes Rockfishes 289447 18187
#> 7 Trachurus symmetricus Jack mackerel 260927 9526
#> 8 Leuroglossus stilbius California smoothtongue 184814 12450
#> 9 Stenobrachius leucopsarus Northern lampfish 178543 12726
#> 10 Triphoturus mexicanus Mexican lampfish 147571 14572Species Distribution
Map the distribution of a common species:
# get Northern Anchovy observations with locations
anchovy <- dbGetQuery(con, "
SELECT
si.latitude as lat,
si.longitude as lon,
SUM(i.tally) as count
FROM ichthyo i
JOIN species sp ON i.species_id = sp.species_id
JOIN net n ON i.net_id = n.net_id
JOIN tow t ON n.tow_id = t.tow_id
JOIN site si ON t.site_id = si.site_id
WHERE sp.scientific_name = 'Engraulis mordax'
GROUP BY si.latitude, si.longitude") |>
filter(!is.na(lon), !is.na(lat)) |>
st_as_sf(coords = c("lon", "lat"), crs = 4326)
mapview(anchovy, cex = "count", layer.name = "Anchovy count")Cruise Timeline
View the temporal coverage of CalCOFI cruises:
# get cruise timeline (date_ym is YYYYMM format)
cruises <- dbGetQuery(con, "
SELECT
cruise_key,
CAST(SUBSTRING(CAST(date_ym AS VARCHAR), 1, 4) AS INTEGER) as year,
CAST(SUBSTRING(CAST(date_ym AS VARCHAR), 5, 2) AS INTEGER) as month,
ship_key
FROM cruise
WHERE date_ym IS NOT NULL
ORDER BY date_ym")
# count cruises by year
cruises |>
count(year) |>
filter(!is.na(year)) |>
ggplot2::ggplot(ggplot2::aes(year, n)) +
ggplot2::geom_col(fill = "steelblue") +
ggplot2::labs(
title = "CalCOFI Cruises by Year",
x = "Year", y = "Number of Cruises") +
ggplot2::theme_minimal()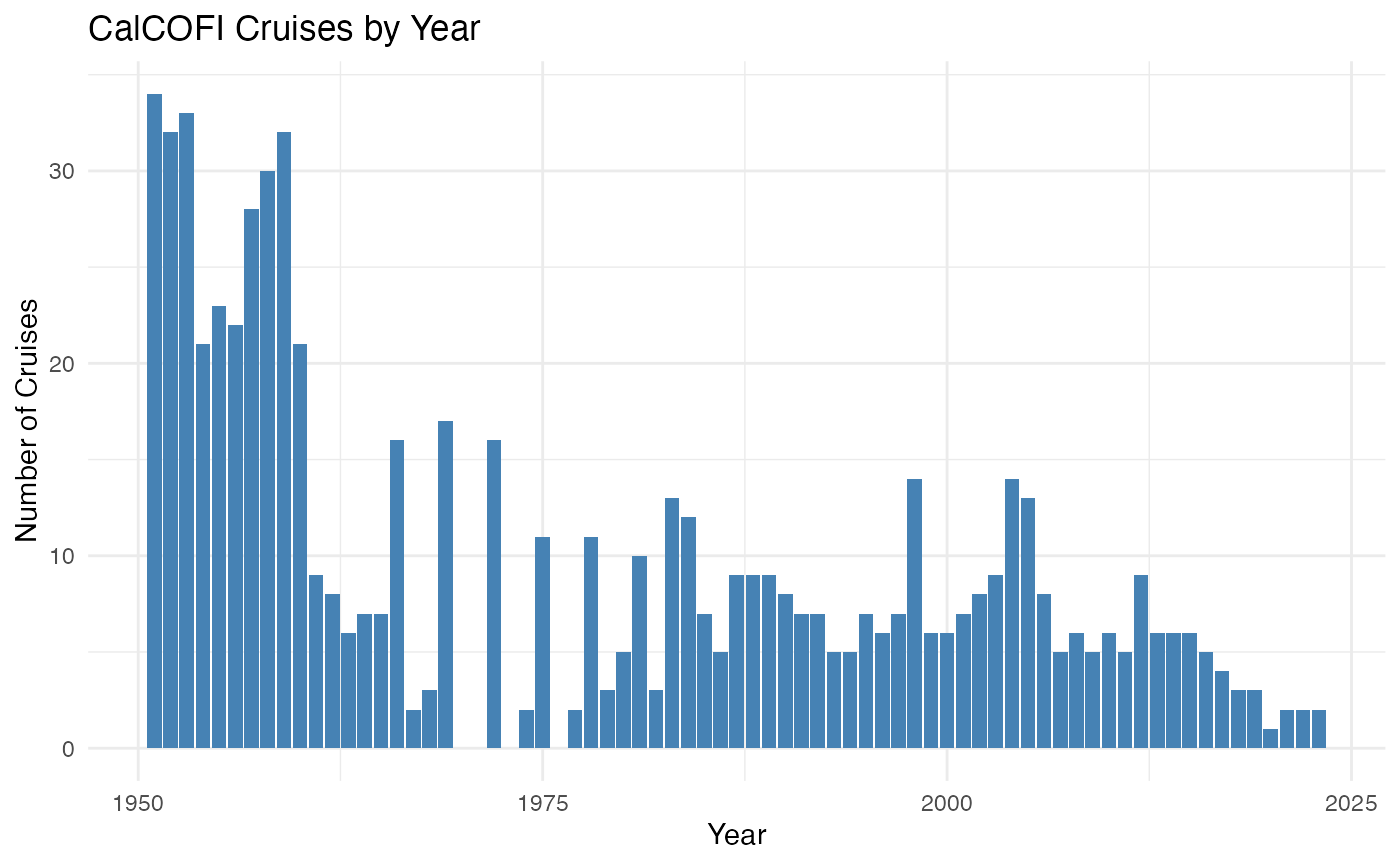
Available Measurement Types
The bottle database includes many oceanographic variables:
# list all measurement types
dbGetQuery(con, "
SELECT measurement_type, description, units
FROM measurement_type
ORDER BY measurement_type") |>
head(20)
#> measurement_type description units
#> 1 alkalinity_rep1 Total alkalinity replicate 1 umol/kg
#> 2 alkalinity_rep2 Total alkalinity replicate 2 umol/kg
#> 3 ammonia Ammonia concentration (QC'd) umol/L
#> 4 barometric_pressure Barometric pressure millibars
#> 5 c14_dark 14C assimilation dark/control bottle mgC/m3/hld
#> 6 c14_mean Mean 14C assimilation of replicates 1 and 2 mgC/m3/hld
#> 7 c14_rep1 14C assimilation replicate 1 mgC/m3/hld
#> 8 c14_rep2 14C assimilation replicate 2 mgC/m3/hld
#> 9 chlorophyll_a Chlorophyll-a measured fluorometrically ug/L
#> 10 cloud_amount Cloud amount in oktas (WMO 2700) oktas
#> 11 cloud_type WMO cloud type code (WMO 0500) <NA>
#> 12 dic_rep1 Dissolved inorganic carbon replicate 1 umol/kg
#> 13 dic_rep2 Dissolved inorganic carbon replicate 2 umol/kg
#> 14 dry_air_temp Dry air temperature from sling psychrometer degC
#> 15 light_pct Light intensity of incubation tubes %
#> 16 nitrate Nitrate concentration umol/L
#> 17 nitrite Nitrite concentration umol/L
#> 18 oxygen_ml_l Dissolved oxygen (QC'd) ml/L
#> 19 oxygen_saturation Oxygen percent saturation %
#> 20 oxygen_umol_kg Dissolved oxygen (QC'd) umol/kgPackage Data Objects
The package also includes pre-loaded spatial data objects for convenience:
-
cc_grid- CalCOFI station grid polygons (sf) -
cc_grid_ctrs- Station centroids (sf) -
cc_grid_zones- Aggregated zone polygons (sf) -
cc_bottle- Sample bottle data for examples (tibble)
These can be used without connecting to the database for quick spatial operations.