---
title: "Ingest NOAA CalCOFI Database"
---
## Overview
**Goal**: Generate the database from source files with workflow scripts
to make updating easier and provenance fully transparent. This allows us
to:
- Rename tables and column names, control data types and use Unicode
encoding for a consistent database ingestion strategy, per Database
naming conventions in [Database – CalCOFI.io
Docs](https://calcofi.io/docs/db.html).
- Differentiate between raw and derived or updated tables and columns.
For instance, the taxonomy for any given species can change over
time, such as lumping or splitting of a given taxa, and by taxonomic
authority (e.g., WoRMS, ITIS or GBIF). These taxonomic identifiers
and the full taxonomic hierarchy should get regularly updated
regardless of source observational data, and can either be updated
in the table directly or joined one-to-one with a seperate table in
a materialized view (so as not to slow down queries with a regular
view).
This workflow processes NOAA CalCOFI database CSV files and outputs
parquet files. The workflow:
1. Reads CSV files from source directory (with GCS archive sync)
2. Loads into local wrangling DuckDB with transformations
3. Restructures primary keys (natural keys + sequential IDs)
4. Creates lookup table and consolidates ichthyo tables
5. Validates data quality and flags issues
6. Exports to parquet files (for later integration into Working DuckLake)
```{mermaid}
%%| label: overview
%%| fig-cap: "Overview diagram of CSV ingestion process into the database."
%%| file: diagrams/ingest_noaa-calcofi-db_overview.mmd
```
See also [5.3 Ingest datasets with documentation – Database – CalCOFI.io Docs](https://calcofi.io/docs/db.html#ingest-datasets-with-documentation) for generic overview of ingestion process.
```{r}
#| label: setup
# devtools::install_local(here::here("../calcofi4db"), force = T)
devtools::load_all(here::here("../calcofi4db"))
devtools::load_all(here::here("../calcofi4r"))
# options(error=NULL)
librarian::shelf(
CalCOFI / calcofi4db,
CalCOFI / calcofi4r,
DBI,
dm,
dplyr,
DT,
fs,
glue,
gargle,
googledrive,
here,
htmltools,
janitor,
jsonlite,
knitr,
listviewer,
litedown,
lubridate,
purrr,
readr,
rlang,
sf,
stringr,
tibble,
tidyr,
units,
uuid,
webshot2,
quiet = T
)
options(readr.show_col_types = F)
options(DT.options = list(scrollX = TRUE))
# define paths
dataset <- "calcofi-db"
provider <- "swfsc.noaa.gov"
dataset_name <- "NOAA CalCOFI Database"
dir_data <- "~/My Drive/projects/calcofi/data-public"
overwrite <- TRUE
dir_parquet <- here(glue("data/parquet/{provider}_{dataset}"))
db_path <- here(glue("data/wrangling/{provider}_{dataset}.duckdb"))
if (overwrite) {
if (file_exists(db_path)) {
file_delete(db_path)
}
if (dir_exists(dir_parquet)) dir_delete(dir_parquet)
}
dir_create(dirname(db_path))
con <- get_duckdb_con(db_path)
load_duckdb_extension(con, "spatial")
# load data using calcofi4db package
# - reads from local Google Drive mount
# - syncs to GCS archive if files changed (creates new timestamped archive)
# - tracks GCS archive path for provenance
d <- read_csv_files(
provider = provider,
dataset = dataset,
dir_data = dir_data,
sync_archive = TRUE,
metadata_dir = here("metadata")
) # workflows/metadata/{provider}/{dataset}/
# show source files summary
message(glue("Loaded {nrow(d$source_files)} tables from {d$paths$dir_csv}"))
message(glue("Total rows: {sum(d$source_files$nrow)}"))
```
## Check for any mismatched tables and fields
```{r}
#| label: data_integrity_checkpoint
#| output: asis
# check data integrity - detects mismatches and controls chunk evaluation
integrity <- check_data_integrity(
d = d,
dataset_name = dataset_name,
halt_on_fail = TRUE
)
# render the pass/fail message
render_integrity_message(integrity)
```
## Show Source Files
```{r}
#| label: source-files
show_source_files(d)
```
## Show CSV Tables and Fields to Ingest
```{r}
#| label: tbls_in
d$d_csv$tables |>
datatable(caption = "Tables to ingest.")
```
```{r}
#| label: flds_in
d$d_csv$fields |>
datatable(caption = "Fields to ingest.")
```
## Show tables and fields redefined
```{r}
#| label: tbls_rd
show_tables_redefine(d)
```
```{r}
#| label: flds_rd
show_fields_redefine(d)
```
## Load Tables into Database
```{r}
#| label: load_tbls_to_db
# use ingest_dataset() which handles:
# - transform_data() for applying redefinitions
# - provenance tracking via gcs_path from read_csv_files()
# - automatic uuid column detection
# - ingest_to_working() for each table
tbl_stats <- ingest_dataset(
con = con,
d = d,
mode = if (overwrite) "replace" else "append",
verbose = TRUE
)
tbl_stats |>
datatable(rownames = FALSE, filter = "top")
```
## Establish Primary Keys
**UUID-first approach**: Source tables (site, tow, net) retain their `*_uuid` columns as
primary unique identifiers. These UUIDs are minted at sea and remain stable throughout
the data lifecycle — even when rows are removed and re-included during QA/QC. Sequential
integer IDs would lose this stability because re-sorting or row additions change the
assignment. Only `cruise` uses a natural key (`cruise_key`) because it has few rows with
easily identifiable attributes (ship + year-month). The `ichthyo` table uses a deterministic
UUID v5 hashed from its composite natural key. Other derived tables without source UUIDs
(lookup, segment) still use sequential integer IDs for convenience.
### Create cruise_key (natural key)
The cruise_key is a natural key in format YYMMKK (2-digit year + 2-digit month + ship_key).
```{r}
#| label: create_cruise_key
# create cruise_key as natural primary key (YYMMKK format)
create_cruise_key(
con,
cruise_tbl = "cruise",
ship_tbl = "ship",
date_col = "date_ym"
)
# verify uniqueness
cruise_keys <- tbl(con, "cruise") |> pull(cruise_key)
stopifnot("cruise_key must be unique" = !any(duplicated(cruise_keys)))
# show sample cruise keys
tbl(con, "cruise") |>
select(cruise_uuid, cruise_key, ship_key, date_ym) |>
head(10) |>
collect() |>
datatable(caption = "Sample cruise_key values (YYMMKK format)")
```
### Propagate cruise_key to child tables
Propagate the natural cruise_key to the site table for convenience in queries and sorting.
The structural foreign key (site.cruise_uuid → cruise.cruise_uuid) comes from the source data.
```{r}
#| label: propagate_cruise_key
# propagate cruise_key from cruise to site (via cruise_uuid)
propagate_natural_key(
con = con,
child_tbl = "site",
parent_tbl = "cruise",
key_col = "cruise_key",
join_col = "cruise_uuid"
)
# verify cruise_key is now in site
tbl(con, "site") |>
select(site_uuid, cruise_uuid, cruise_key, orderocc) |>
head(10) |>
collect() |>
datatable(caption = "Sample site rows with cruise_key")
```
## Create Lookup Table
Create unified lookup table from vocabularies for egg stages, larva stages, and tow types.
```{r}
#| label: create_lookup_table
# egg stage vocabulary (Moser & Ahlstrom, 1985)
egg_stage_vocab <- tibble(
stage_int = 1:11,
stage_description = c(
"egg, stage 1 of 11 (Moser & Ahlstrom, 1985)",
"egg, stage 2 of 11 (Moser & Ahlstrom, 1985)",
"egg, stage 3 of 11 (Moser & Ahlstrom, 1985)",
"egg, stage 4 of 11 (Moser & Ahlstrom, 1985)",
"egg, stage 5 of 11 (Moser & Ahlstrom, 1985)",
"egg, stage 6 of 11 (Moser & Ahlstrom, 1985)",
"egg, stage 7 of 11 (Moser & Ahlstrom, 1985)",
"egg, stage 8 of 11 (Moser & Ahlstrom, 1985)",
"egg, stage 9 of 11 (Moser & Ahlstrom, 1985)",
"egg, stage 10 of 11 (Moser & Ahlstrom, 1985)",
"egg, stage 11 of 11 (Moser & Ahlstrom, 1985)"
)
)
# larva stage vocabulary
larva_stage_vocab <- tibble(
stage_int = 1:5,
stage_txt = c("YOLK", "PREF", "FLEX", "POST", "TRNS"),
stage_description = c(
"larva, yolk sac",
"larva, preflexion",
"larva, flexion",
"larva, postflexion",
"larva, transformation"
)
)
# tow type vocabulary (from tow_type table)
tow_type_vocab <- tbl(con, "tow_type") |>
collect() |>
mutate(
lookup_num = row_number(),
lookup_chr = tow_type_key,
description = description
) |>
select(lookup_num, lookup_chr, description)
# create unified lookup table
create_lookup_table(
con = con,
egg_stage_vocab = egg_stage_vocab,
larva_stage_vocab = larva_stage_vocab,
tow_type_vocab = tow_type_vocab
)
# show lookup table
tbl(con, "lookup") |>
collect() |>
datatable(caption = "Lookup table with vocabularies")
```
## Consolidate to Tidy Ichthyo Table
Consolidate 5 ichthyoplankton tables (egg, eggstage, larva, larvastage, larvasize) into
a single tidy table.
```{mermaid}
%%| label: consolidate_ichthyo_diagram
%%| fig-cap: "Consolidate 5 ichthyoplankton tables into a single tidy ichthyo table with lookup."
erDiagram
egg {
uuid net_uuid FK
int species_id FK
int tally
}
egg_stage {
uuid net_uuid FK
int species_id FK
int stage
int tally
}
larva {
uuid net_uuid FK
int species_id FK
int tally
}
larva_stage {
uuid net_uuid FK
int species_id FK
str stage
int tally
}
larva_size {
uuid net_uuid FK
int species_id FK
dbl length_mm
int tally
}
ichthyo {
uuid ichthyo_uuid PK
uuid net_uuid FK
int species_id FK
str life_stage
str measurement_type
dbl measurement_value
int tally
}
lookup {
int lookup_id PK
str category
int lookup_num
str lookup_chr
str description
}
egg ||--|{ ichthyo : "life_stage = egg"
egg_stage ||--|{ ichthyo : "life_stage = egg, type = stage"
larva ||--|{ ichthyo : "life_stage = larva"
larva_stage ||--|{ ichthyo : "life_stage = larva, type = stage"
larva_size ||--|{ ichthyo : "life_stage = larva, type = size"
ichthyo }o--|| lookup : "measurement_value"
```
```{r}
#| label: consolidate_ichthyo
message("Consolidating ichthyoplankton tables...")
# consolidate all ichthyo tables (keeps net_uuid as FK to net table)
consolidate_ichthyo_tables(
con = con,
output_tbl = "ichthyo",
larva_stage_vocab = larva_stage_vocab
)
# assign ichthyo_uuid — deterministic UUID v5 from composite natural key
assign_deterministic_uuids(
con = con,
table_name = "ichthyo",
id_col = "ichthyo_uuid",
key_cols = c(
"net_uuid",
"species_id",
"life_stage",
"measurement_type",
"measurement_value")
)
# show sample rows
tbl(con, "ichthyo") |>
head(20) |>
collect() |>
datatable(caption = "Sample ichthyo table rows (tidy format)")
```
```{r}
#| label: ichthyo_summary
# summarize ichthyo table
ichthyo_summary <- tbl(con, "ichthyo") |>
group_by(life_stage, measurement_type) |>
summarize(
n_rows = n(),
n_species = n_distinct(species_id),
total_tally = sum(tally, na.rm = TRUE),
.groups = "drop"
) |>
collect()
ichthyo_summary |>
arrange(life_stage, measurement_type) |>
datatable(
caption = HTML(mark(
"The `ichthyo` table summary by life_stage and measurement_type"
))
) |>
formatCurrency(
columns = c("n_rows", "n_species", "total_tally"),
currency = "",
digits = 0
)
```
## Data Quality Improvements
This section applies data corrections and validates referential integrity.
### Data Corrections
Apply known data corrections identified by data managers.
```{r}
#| label: apply_corrections
# apply data corrections
apply_data_corrections(con, verbose = TRUE)
```
### Validate Referential Integrity
Run validation checks and flag invalid rows for review.
```{r}
#| label: validate_integrity
# ensure flagged directory exists
dir_flagged <- here("data/flagged")
if (!dir.exists(dir_flagged)) {
dir.create(dir_flagged, recursive = TRUE)
}
# validate egg stages (must be 1-11)
invalid_egg_stages <- validate_egg_stages(con, "egg_stage", "stage")
invalid_egg_stages_csv <- file.path(dir_flagged, "invalid_egg_stages.csv")
invalid_egg_stages_desc <- "Egg stage values NOT 1 to 11 (ie, not in Moser & Ahlstrom 1985 vocab)"
if (nrow(invalid_egg_stages) > 0) {
flag_invalid_rows(
invalid_rows = invalid_egg_stages,
output_path = invalid_egg_stages_csv,
description = invalid_egg_stages_desc
)
}
show_flagged_file(
invalid_egg_stages,
invalid_egg_stages_csv,
invalid_egg_stages_desc
)
# define validation checks
validations <- list(
list(
type = "fk",
data_tbl = "ichthyo",
col = "species_id",
ref_tbl = "species",
ref_col = "species_id",
output_file = "orphan_species.csv",
description = "Species IDs not found in species table"
),
list(
type = "fk",
data_tbl = "ichthyo",
col = "net_uuid",
ref_tbl = "net",
ref_col = "net_uuid",
output_file = "orphan_nets.csv",
description = "Net UUIDs not found in net table"
)
)
# run validations
validation_results <- validate_dataset(
con = con,
validations = validations,
output_dir = dir_flagged
)
# show validation summary with GitHub links
show_validation_results(validation_results)
```
```{r}
#| label: delete_flagged
# optionally delete flagged rows (dry run first)
if (validation_results$total_flagged > 0) {
message(glue("Found {validation_results$total_flagged} invalid rows"))
# dry run to see what would be deleted
delete_stats <- delete_flagged_rows(
con = con,
validation_results = validation_results,
dry_run = TRUE
)
delete_stats |> datatable(caption = "Rows to be deleted (dry run)")
# uncomment to actually delete:
delete_flagged_rows(con, validation_results, dry_run = FALSE)
}
```
### Drop Deprecated Tables
The source tables have been consolidated into `ichthyo` (tidy format) and `lookup`
(vocabularies). Drop these before creating the schema diagram.
Note: `*_uuid` columns are retained as primary unique identifiers (see rationale
in "Establish Primary Keys" above).
```{r}
#| label: drop_deprecated
# tables consolidated into ichthyo
deprecated_ichthyo <- c(
"egg",
"egg_stage",
"larva",
"larva_stage",
"larva_size"
)
# tables consolidated into lookup
deprecated_lookup <- c("tow_type")
# all deprecated tables
deprecated_tables <- c(deprecated_ichthyo, deprecated_lookup)
# drop each deprecated table
for (tbl in deprecated_tables) {
if (tbl %in% DBI::dbListTables(con)) {
DBI::dbExecute(con, glue("DROP TABLE {tbl}"))
message(glue("Dropped deprecated table: {tbl}"))
}
}
message(glue(
"\nRemaining tables: {paste(sort(DBI::dbListTables(con)), collapse = ', ')}"
))
```
## Schema Documentation
```{r}
#| label: dm_tbls
tbls <- dbListTables(con) |> sort()
# dm object for visualization (keys learned from data, not DB constraints)
dm_dev <- dm_from_con(con, table_names = tbls, learn_keys = FALSE)
dm_draw(dm_dev, view_type = "all")
```
### Primary Key Strategy
**UUID-first**: Source tables use UUIDs minted at sea as primary identifiers.
Only `cruise` uses a natural key because it has few, easily identifiable rows.
Derived tables (lookup, segment) use sequential integer IDs since they
have no source UUID. The `ichthyo` table uses a deterministic UUID v5 hashed
from its composite natural key.
| Table | Primary Key | Type |
|-------|-------------|------|
| `cruise` | `cruise_key` | Natural key (YYMMKK); also retains `cruise_uuid` from source |
| `ship` | `ship_key` | Natural key (2-letter) |
| `site` | `site_uuid` | Source UUID (minted at sea) |
| `tow` | `tow_uuid` | Source UUID (minted at sea) |
| `net` | `net_uuid` | Source UUID (minted at sea) |
| `species` | `species_id` | Natural from source |
| `ichthyo` | `ichthyo_uuid` | Deterministic UUID v5 (from net_uuid, species_id, life_stage, measurement_type, measurement_value) |
| `lookup` | `lookup_id` | Sequential (derived table, sorted by lookup_type, lookup_num) |
### Foreign Key Relationships
DuckDB doesn't support `ALTER TABLE ADD FOREIGN KEY`. We use the dm package for documentation and visualization only.
**Foreign Key Relationships:**
```
ship.ship_key (PK)
↓
cruise.cruise_key (PK) ←── cruise.ship_key (FK)
↓ cruise.cruise_uuid (unique, source)
site.site_uuid (PK) ←── site.cruise_uuid (FK → cruise.cruise_uuid)
↓ site.cruise_key (denormalized, for queries)
tow.tow_uuid (PK) ←── tow.site_uuid (FK → site.site_uuid)
↓ tow.tow_type_key → lookup (lookup_type='tow_type')
net.net_uuid (PK) ←── net.tow_uuid (FK → tow.tow_uuid)
↓
ichthyo.ichthyo_uuid (PK) ←── ichthyo.net_uuid (FK → net.net_uuid)
ichthyo.species_id (FK) → species.species_id
```
```{r}
#| label: dm_fk
# get tables (excluding old ichthyo source tables that are now consolidated)
tbls <- sort(DBI::dbListTables(con))
# refresh dm after index creation
dm_dev <- dm_from_con(con, table_names = tbls, learn_keys = FALSE)
# define relationships in dm (in-memory for visualization only)
# uses UUID PKs for source tables, sequential IDs for derived tables
add_cc_keys <- function(dm) {
dm |>
dm_add_pk(cruise, cruise_key) |>
dm_add_pk(ship, ship_key) |>
dm_add_pk(site, site_uuid) |>
dm_add_pk(tow, tow_uuid) |>
dm_add_pk(net, net_uuid) |>
dm_add_pk(species, species_id) |>
dm_add_pk(ichthyo, ichthyo_uuid) |>
dm_add_pk(lookup, lookup_id) |>
dm_add_fk(ichthyo, net_uuid, net) |>
dm_add_fk(ichthyo, species_id, species) |>
dm_add_fk(net, tow_uuid, tow) |>
dm_add_fk(tow, site_uuid, site) |>
dm_add_fk(site, cruise_key, cruise) |>
dm_add_fk(cruise, ship_key, ship)
}
dm_dev_fk <- dm_dev |>
add_cc_keys()
dm_draw(dm_dev_fk, view_type = "all")
```
## Add Spatial
### Add `site.geom`
```{r}
#| label: mk_site_pts
# add geometry column using DuckDB spatial
# note: DuckDB spatial doesn't track SRID metadata (unlike PostGIS)
# all geometries assumed WGS84 (EPSG:4326) by convention
add_point_geom(con, "site", lon_col = "longitude", lat_col = "latitude")
```
### Fix calcofi4r `grid`
Problems with
[calcofi4r::`cc_grid`](https://calcofi.io/calcofi4r/articles/calcofi4r.html):
- uses old station (line, position) vs newer site (line, station)
- `sta_lin`, `sta_pos`: integer, so drops necessary decimal that is found in `sta_key`
- `sta_lin == 90, sta_pos == 120` repeats for:
- `sta_pattern` == 'historical' (`sta_dpos` == 20); and
- `sta_pattern` == 'standard' (`sta_dpos` == 10)
```{r}
#| label: mk_grid_v2
librarian::shelf(
calcofi4r,
mapview,
quiet = T
)
cc_grid_v2 <- calcofi4r::cc_grid |>
select(
sta_key,
shore = sta_shore,
pattern = sta_pattern,
spacing = sta_dpos
) |>
separate_wider_delim(
sta_key,
",",
names = c("line", "station"),
cols_remove = F
) |>
mutate(
line = as.double(line),
station = as.double(station),
grid_key = ifelse(
pattern == "historical",
glue("st{station}-ln{line}_hist"),
glue("st{station}-ln{line}")
),
zone = glue("{shore}-{pattern}")
) |>
relocate(grid_key, station) |>
st_as_sf() |>
mutate(
area_km2 = st_area(geom) |>
set_units(km^2) |>
as.numeric()
)
cc_grid_ctrs_v2 <- calcofi4r::cc_grid_ctrs |>
select(sta_key, pattern = sta_pattern) |>
left_join(
cc_grid_v2 |>
st_drop_geometry(),
by = c("sta_key", "pattern")
) |>
select(-sta_key) |>
relocate(grid_key)
cc_grid_v2 <- cc_grid_v2 |>
select(-sta_key)
cc_grid_v2 |>
st_drop_geometry() |>
datatable()
mapview(cc_grid_v2, zcol = "zone") +
mapview(cc_grid_ctrs_v2, cex = 1)
```
```{r}
#| label: grid_to_db
grid <- cc_grid_v2 |>
as.data.frame() |>
left_join(
cc_grid_ctrs_v2 |>
as.data.frame() |>
select(grid_key, geom_ctr = geom),
by = "grid_key"
) |>
st_as_sf(sf_column_name = "geom")
# convert sf geometry to WKB for DuckDB
grid_df <- grid |>
mutate(
geom_wkb = sf::st_as_binary(geom, hex = TRUE),
geom_ctr_wkb = sf::st_as_binary(geom_ctr, hex = TRUE)
) |>
sf::st_drop_geometry() |>
select(-geom_ctr)
# write to DuckDB
dbWriteTable(con, "grid", grid_df, overwrite = TRUE)
# convert WKB back to geometry
dbExecute(con, "ALTER TABLE grid ADD COLUMN geom GEOMETRY")
dbExecute(con, "UPDATE grid SET geom = ST_GeomFromHEXWKB(geom_wkb)")
dbExecute(con, "ALTER TABLE grid DROP COLUMN geom_wkb")
dbExecute(con, "ALTER TABLE grid ADD COLUMN geom_ctr GEOMETRY")
dbExecute(con, "UPDATE grid SET geom_ctr = ST_GeomFromHEXWKB(geom_ctr_wkb)")
dbExecute(con, "ALTER TABLE grid DROP COLUMN geom_ctr_wkb")
message("Grid table created with geometry columns")
```
### Update `site.grid_key`
```{r}
#| label: update_site_from_grid
grid_stats <- assign_grid_key(con, "site")
grid_stats |> datatable()
```
### Add `segment`: line segments between consecutive sites
```{r}
#| label: mk_segment
segment <- tbl(con, "site") |>
select(cruise_key, orderocc, site_uuid, lon = longitude, lat = latitude) |>
left_join(
tbl(con, "tow") |>
select(site_uuid, time_start),
by = "site_uuid"
) |>
group_by(
cruise_key,
orderocc,
site_uuid,
lon,
lat
) |>
summarize(
time_beg = min(time_start, na.rm = T),
time_end = max(time_start, na.rm = T),
.groups = "drop"
) |>
collect()
segment <- segment |>
arrange(cruise_key, orderocc, time_beg) |>
group_by(cruise_key) |>
mutate(
site_uuid_beg = lag(site_uuid),
lon_beg = lag(lon),
lat_beg = lag(lat),
time_beg = lag(time_beg)
) |>
ungroup() |>
filter(!is.na(lon_beg), !is.na(lat_beg)) |>
mutate(
m = pmap(
list(lon_beg, lat_beg, lon, lat),
\(x1, y1, x2, y2) {
matrix(c(x1, y1, x2, y2), nrow = 2, byrow = T)
}
),
geom = map(m, st_linestring)
) |>
select(
cruise_key,
site_uuid_beg,
site_uuid_end = site_uuid,
lon_beg,
lat_beg,
lon_end = lon,
lat_end = lat,
time_beg,
time_end,
geom
) |>
st_as_sf(
sf_column_name = "geom",
crs = 4326
) |>
mutate(
time_hr = as.numeric(difftime(time_end, time_beg, units = "hours")),
length_km = st_length(geom) |>
set_units(km) |>
as.numeric(),
km_per_hr = length_km / time_hr
)
# convert to WKB and write to DuckDB
segment_df <- segment |>
mutate(geom_wkb = sf::st_as_binary(geom, hex = TRUE)) |>
sf::st_drop_geometry()
dbWriteTable(con, "segment", segment_df, overwrite = TRUE)
dbExecute(con, "ALTER TABLE segment ADD COLUMN geom GEOMETRY")
dbExecute(con, "UPDATE segment SET geom = ST_GeomFromHEXWKB(geom_wkb)")
dbExecute(con, "ALTER TABLE segment DROP COLUMN geom_wkb")
# assign segment_id sorted by time_beg
assign_sequential_ids(
con = con,
table_name = "segment",
id_col = "segment_id",
sort_cols = c("time_beg")
)
message("Segment table created")
```
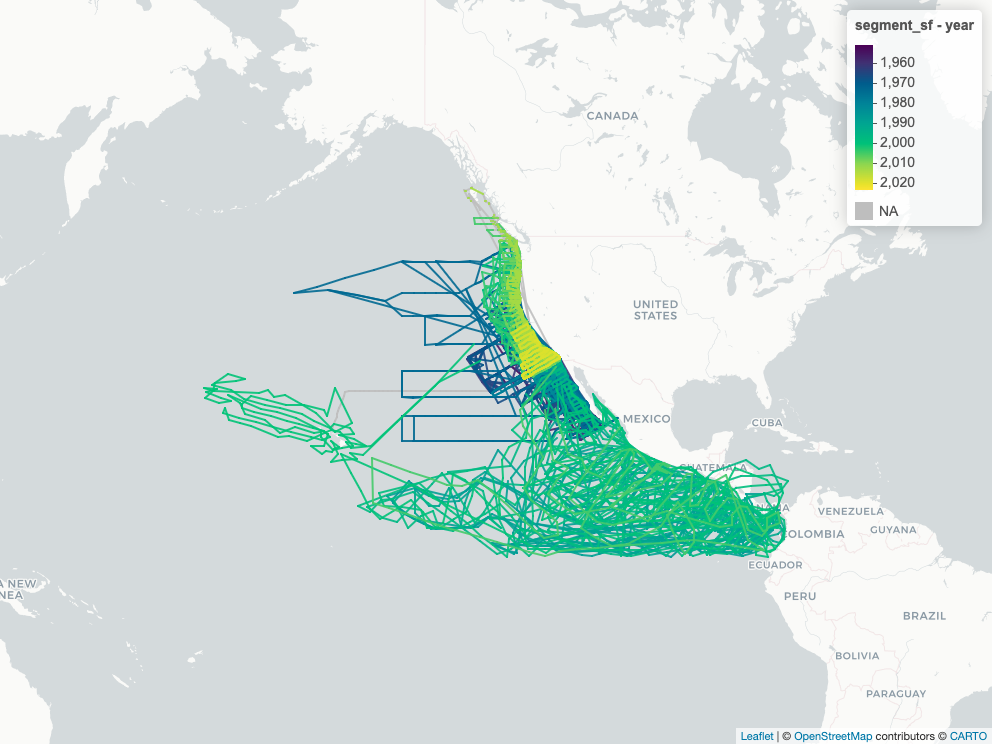
```{r}
#| label: view_segment
# slowish, so use cached figure
map_segment_png <- here(glue("figures/{provider}_{dataset}_segment_map.png"))
if (!file_exists(map_segment_png)) {
segment_sf <- cc_read_sf(con, "segment") |>
mutate(year = year(time_beg))
m <- mapView(segment_sf, zcol = "year")
mapshot2(m, file = map_segment_png)
}
htmltools::img(
src = map_segment_png |> str_replace(here(), "."),
width = "600px"
)
```
## Report
```{r}
#| label: show_latest
all_tbls <- DBI::dbListTables(con)
add_cc_spatial_keys <- function(dm) {
dm |>
dm_add_pk(grid, grid_key) |>
dm_add_pk(segment, segment_id) |>
dm_add_fk(site, grid_key, grid) |>
dm_add_fk(segment, cruise_key, cruise)
}
dm_final <- dm_from_con(con, table_names = all_tbls, learn_keys = FALSE) |>
add_cc_keys() |>
add_cc_spatial_keys() |>
dm_set_colors(lightgreen = c(segment, grid))
dm_draw(dm_final, view_type = "all")
```
```{r}
#| label: effort_stats
d_eff <- tbl(con, "segment") |>
mutate(
year = year(time_beg)
) |>
group_by(year) |>
summarize(
time_hr = sum(time_hr, na.rm = T),
length_km = sum(length_km, na.rm = T)
) |>
collect()
total_hours <- sum(d_eff$time_hr, na.rm = T)
total_km <- sum(d_eff$length_km, na.rm = T)
fmt <- function(x, ...) format(x, big.mark = ",", ...)
message(glue(
"Total effort: {fmt(round(total_hours))} hours ({fmt(round(total_hours/24))} days, {fmt(round(total_hours/24/365, 1))} years)"
))
message(glue("Total distance: {fmt(round(total_km))} km"))
```
## Validate Local Database
Validate data quality in the local wrangling database before exporting to parquet.
The parquet outputs from this workflow can later be used to update the Working DuckLake.
```{r}
#| label: validate
# validate data quality
validation <- validate_for_release(con)
if (validation$passed) {
message("Validation passed!")
if (nrow(validation$checks) > 0) {
validation$checks |>
filter(status != "pass") |>
datatable(caption = "Validation Warnings")
}
} else {
cat("Validation FAILED:\n")
cat(paste("-", validation$errors, collapse = "\n"))
}
```
## Enforce Column Types
Force integer/smallint types on columns that R's `numeric` mapped to `DOUBLE`
during `dbWriteTable()`. Uses `flds_redefine.csv` (`type_new`) as the source of
truth for source-table columns, plus explicit overrides for derived-table columns.
```{r}
#| label: enforce_types
type_changes <- enforce_column_types(
con = con,
d_flds_rd = d$d_flds_rd,
type_overrides = list(
ichthyo.ichthyo_uuid = "UUID",
ichthyo.species_id = "SMALLINT",
ichthyo.tally = "INTEGER",
lookup.lookup_id = "INTEGER",
lookup.lookup_num = "INTEGER",
segment.segment_id = "INTEGER"
),
tables = dbListTables(con),
verbose = TRUE
)
if (nrow(type_changes) > 0) {
type_changes |>
datatable(caption = "Column type changes applied")
}
```
## Data Preview
Preview first and last rows of each table before writing parquet outputs.
```{r}
#| label: preview_tables
#| results: asis
preview_tables(
con,
c(
"cruise",
"ship",
"site",
"tow",
"net",
"species",
"ichthyo",
"lookup",
"grid",
"segment"
)
)
```
## Write Parquet Outputs
Export tables to parquet files for downstream use.
```{r}
#| label: write_parquet
# write parquet files with manifest
parquet_stats <- write_parquet_outputs(
con = con,
output_dir = dir_parquet,
tables = dbListTables(con),
strip_provenance = F
)
parquet_stats |>
mutate(file = basename(path)) |>
select(-path) |>
datatable(caption = "Parquet export statistics")
```
## Write Metadata
Build `metadata.json` sidecar file documenting all tables and columns in parquet outputs. DuckDB `COMMENT ON` does not propagate to parquet, so this provides the metadata externally.
```{r}
#| label: write_metadata
metadata_path <- build_metadata_json(
con = con,
d_tbls_rd = d$d_tbls_rd,
d_flds_rd = d$d_flds_rd,
metadata_derived_csv = here(
"metadata/swfsc.noaa.gov/calcofi-db/metadata_derived.csv"
),
output_dir = dir_parquet,
tables = dbListTables(con),
set_comments = TRUE,
provider = provider,
dataset = dataset,
workflow_url = glue(
"https://calcofi.io/workflows/ingest_swfsc.noaa.gov_calcofi-db.html"
)
)
# show metadata summary
metadata <- jsonlite::fromJSON(metadata_path)
tibble(
table = names(metadata$tables),
n_cols = map_int(
names(metadata$tables),
~ sum(grepl(glue("^{.x}\\."), names(metadata$columns)))
),
name_long = map_chr(metadata$tables, ~ .x$name_long)
) |>
datatable(caption = "Table metadata summary")
```
```{r}
#| label: show_metadata_json
listviewer::jsonedit(
jsonlite::fromJSON(metadata_path, simplifyVector = FALSE),
mode = "view"
)
```
## Upload to GCS Archive
Upload parquet files, manifest, and metadata sidecar to `gs://calcofi-db/ingest/{provider}_{dataset}/`.
```{r}
#| label: upload_gcs
gcs_ingest_prefix <- glue("ingest/{provider}_{dataset}")
gcs_bucket <- "calcofi-db"
# sync to GCS — only uploads new or changed files
sync_results <- sync_to_gcs(
local_dir = dir_parquet,
gcs_prefix = gcs_ingest_prefix,
bucket = gcs_bucket)
```
## Cleanup
```{r}
#| label: cleanup
# close local wrangling database connection
close_duckdb(con)
message("Local wrangling database connection closed")
# note: parquet outputs are in data/parquet/swfsc.noaa.gov_calcofi-db/
# these can be used to update the Working DuckLake in a separate workflow
message(glue("Parquet outputs written to: {dir_parquet}"))
message(glue("GCS outputs at: gs://{gcs_bucket}/{gcs_ingest_prefix}/"))
```
## TODO
- [ ] Review documentation for more comment descriptions at [Data \> Data Formats \| CalCOFI.org](https://calcofi.com/index.php?option=com_content&view=category&id=73&Itemid=993)
::: {.callout-caution collapse="true"}
## Session Info
```{r session_info}
devtools::session_info()
```
:::