Code
source(here::here("../apps_dev/libs/db.R")) # con: database connection
librarian::shelf(
calcofi/calcofi4r,
dm, DT, glue, purrr, stringr, tidyr)Goal: Systematically evaluate and clean-up the database, including renaming tables and columns, and documenting relationships, per naming conventions being established in CalCOFI/docs: Database.
source(here::here("../apps_dev/libs/db.R")) # con: database connection
librarian::shelf(
calcofi/calcofi4r,
dm, DT, glue, purrr, stringr, tidyr)Let’s evaluate tables that are in transition, i.e. containing in the table name: “old”, “new”, “backup”, “temp”, “temp”, or “test”.
d_tbls <- tibble(
table = dbListTables(con)) |>
mutate(
subset = case_when(
str_detect(table, "old") ~ "old",
str_detect(table, "new") ~ "new",
str_detect(table, "backup") ~ "backup",
str_detect(table, "temp|tmp") ~ "temp",
str_detect(table, "test") ~ "test",
TRUE ~ "base")) |>
arrange(subset, table)
table(d_tbls$subset)
backup base new old temp test
3 68 5 4 3 1 d_tbls |>
filter(subset != "base")# A tibble: 16 × 2
table subset
<chr> <chr>
1 egg_counts_backup backup
2 larvae_counts_backup backup
3 nets_backup backup
4 newnet2ctdcast new
5 scrippscast_new new
6 species_codes_new new
7 species_new new
8 tow_types_new new
9 egg_counts_old old
10 larvae_counts_old old
11 stations_old old
12 tows_old old
13 net_uuids_tmp temp
14 tempnet2ctd temp
15 tmp_out temp
16 test_rast test tbls_other <- d_tbls |>
filter(subset != "base") |>
pull(table)
cc_db_catalog(tbls_other)TODO: Evaluate these tables and decide what to do with them, i.e. rename or drop.tbls_base <- d_tbls |>
filter(subset == "base") |>
pull(table) %>%
c(., "species_codes_new") |>
sort()
tibble(
table = tbls_base)# A tibble: 69 × 1
table
<chr>
1 aoi_fed_sanctuaries
2 core_stations
3 cruises
4 cruises_uuids
5 ctd_bottles
6 ctd_cast_bottles
7 ctd_casts
8 ctd_dic
9 ctd_profiles
10 dataset_keywords
# ℹ 59 more rowscc_db_catalog(tables = tbls_base)Let’s evaluate tables that are taxonomic in nature, including “species”, “taxa”.
d_tbls <- tibble(
table = dbListTables(con)) |>
mutate(
subset = case_when(
str_detect(table, "species") ~ "species",
str_detect(table, "taxa") ~ "taxa")) |>
filter(
!is.na(subset)) |>
arrange(subset, table) |>
relocate(subset)
datatable(d_tbls)cc_db_catalog(d_tbls$table)Let’s look at redundancy of tables labeled with “uuids”.
tbls_uuids <- str_subset(dbListTables(con), "uuids")
tbls_pfx <- str_replace(tbls_uuids, "(.*)_uuids.*", "\\1")
d_tbls <- tibble(
table = dbListTables(con)) |>
mutate(
subset = case_when(
table %in% tbls_uuids ~ "uuids",
table %in% tbls_pfx ~ "prefix")) |>
filter(
!is.na(subset)) |>
arrange(table, subset)
datatable(d_tbls)cc_db_catalog(d_tbls$table)Generalize from “species” to “taxa” (or “taxon” singular), since not all species_codes are at taxonomic level of species. OR operate from understanding that species could refer to other taxon ranks.
Follow conventions at calcofi.io/docs: Database.
See CalCOFI/workflows: modify_db.R for previous SQL commands, including renames, and dates.
TODO: Consider other changes and how to document. For instance, perhaps track changes in the pg_dump of only data definition language (DDL) for the whole database (i.e., CREATE TABLE SQL statements), and see git differences over time.
pg_dump -U user_name -h host database -s -t table_or_view_names -f table_or_view_names.sql# TODO: species_codes -> taxa
q("ALTER TABLE species_codes RENAME TO taxa")
# TODO: species.spccode -> sp_key
q("ALTER TABLE taxa RENAME COLUMN spccode TO taxa_key")dm R package)"keys_only"con_dm <- dm_from_con(
con,
table_names = tbls_base,
learn_keys = T)
con_dm── Table source ────────────────────────────────────────────────────────────────
src: postgres [admin@localhost:5432/gis]
── Metadata ────────────────────────────────────────────────────────────────────
Tables: `aoi_fed_sanctuaries`, `core_stations`, `cruises`, `cruises_uuids`, `ctd_bottles`, … (69 total)
Columns: 642
Primary keys: 14
Foreign keys: 5dm_draw(con_dm, view_type = "keys_only")"all"This is too much.
# TODO: make SVG zoomable
dm_draw(con_dm, view_type = "all")dm_get_all_uks(con_dm)# A tibble: 14 × 3
table uk_col kind
<chr> <keys> <chr>
1 cruises cruise_id PK
2 ctd_bottles btl_cnt PK
3 ctd_casts cast_count PK
4 grd_a rid PK
5 grd_gcs rid PK
6 grd_mer rid PK
7 nets netid PK
8 r_mer100km rid PK
9 r_sta_cnt rid PK
10 spatial_ref_sys srid PK
11 stations stationid PK
12 tows towid PK
13 users email PK
14 z_idw rid PK # candidate primary keys
dm_enum_pk_candidates(con_dm, "species_codes_new")# A tibble: 6 × 3
columns candidate why
<keys> <lgl> <chr>
1 id TRUE ""
2 spccode TRUE ""
3 scientific_name TRUE ""
4 itis_tsn TRUE ""
5 common_name FALSE "has duplicate values: NULL (366), Dreamers …
6 taxon_rank FALSE "has 34 missing values, and duplicate values: Speci…# candidate foreign keys
try(
dm_enum_fk_candidates(con_dm, "species_codes_new", "species_groups"))Error in abort_ref_tbl_has_no_pk(ref_table_name) :
ref_table `species_groups` needs a primary key first. Use `dm_enum_pk_candidates()` to find appropriate columns and `dm_add_pk()` to define a primary key.# install.packages("DiagrammeRsvg")
dm_gui(dm = con_dm)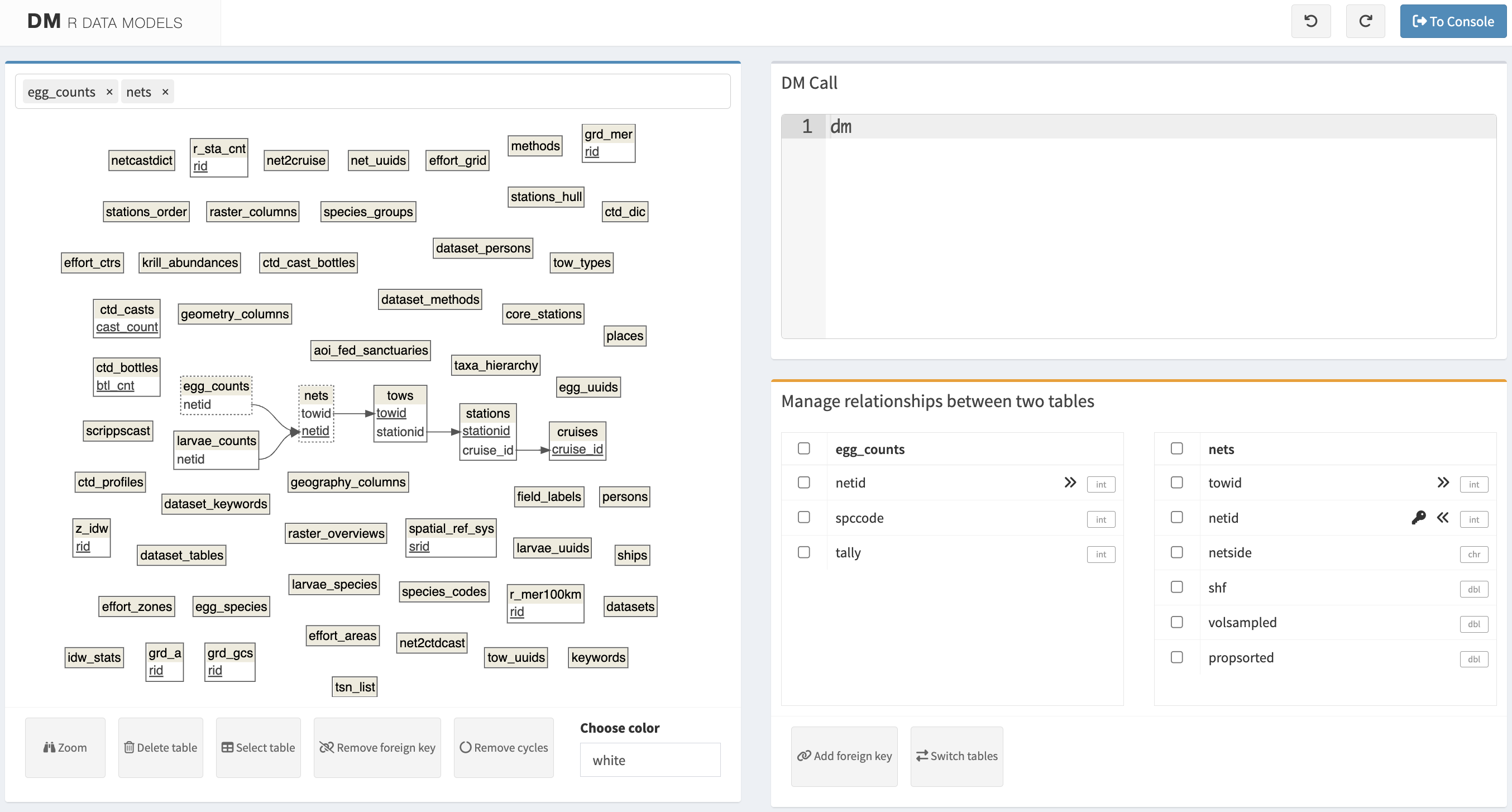
dm::dm_gui() Shiny app after clicking on two tables. Haven’t tried editing relationships yet.