---
title: "Ingest Bottle Database"
---
## Overview
**Source**: [Bottle Database | CalCOFI.org](https://calcofi.org/data/oceanographic-data/bottle-database/)
- Download: <https://calcofi.org/downloads/database/CalCOFI_Database_194903-202105_csv_16October2023.zip>
**Goal**: Ingest bottle database from source CSV files into a local
wrangling DuckDB, then export as parquet files for downstream use.
**Two source tables**:
- `194903-202105_Cast.csv` (61 fields) — station-level: cruise,
location, meteorology, integrated measurements
- `194903-202105_Bottle.csv` (62 fields) — sample-level: depth,
temperature, salinity, O₂, nutrients, pigments, C14, pH
**Coverage**: 1949-03 to 2021-05 (72 years)
**Join key**: `cast_id` (in both tables; source column `Cst_Cnt`)
This workflow processes CalCOFI bottle database CSV files and outputs
parquet files. The workflow:
1. Reads CSV files from source directory (with GCS archive sync)
2. Loads into local wrangling DuckDB with column renames
3. Applies ship name/code corrections
4. Consolidates temporal columns into `datetime_utc`, drops derivable columns, creates `casts_derived` VIEW
5. Pivots bottle measurements into long-format `bottle_measurement` table
6. Pivots cast environmental conditions into `cast_condition` table
7. Loads `measurement_type` reference table
8. Validates primary keys and foreign key integrity
9. Adds spatial geometry and grid assignments
10. Exports to parquet files (for later integration into Working DuckLake)
```{r}
#| label: setup
# devtools::install_local(here::here("../calcofi4db"), force = T)
devtools::load_all(here::here("../calcofi4db"))
devtools::load_all(here::here("../calcofi4r"))
librarian::shelf(
CalCOFI/calcofi4db,
CalCOFI/calcofi4r,
DBI, dm, dplyr, DT, fs, glue, gargle, googledrive, here, htmltools,
janitor, jsonlite, knitr, listviewer, litedown, lubridate, mapview,
purrr, readr, rlang, sf, stringr, tibble, tidyr, units, webshot2,
quiet = T)
options(readr.show_col_types = F)
options(DT.options = list(scrollX = TRUE))
# define paths
provider <- "calcofi.org"
dataset <- "bottle-database"
dataset_name <- "CalCOFI Bottle Database"
dir_data <- "~/My Drive/projects/calcofi/data-public"
subdir <- "CalCOFI_Database_194903-202105_csv_16October2023"
overwrite <- TRUE
dir_parquet <- here(glue("data/parquet/{provider}_{dataset}"))
db_path <- here(glue("data/wrangling/{provider}_{dataset}.duckdb"))
if (overwrite){
if (file_exists(db_path)) file_delete(db_path)
if (dir_exists(dir_parquet)) dir_delete(dir_parquet)
}
dir_create(dirname(db_path))
con <- get_duckdb_con(db_path)
load_duckdb_extension(con, "spatial")
# load data using calcofi4db package
# - reads from local Google Drive mount
# - syncs to GCS archive if files changed (creates new timestamped archive)
# - tracks GCS archive path for provenance
dir_dataset <- glue("{dir_data}/{provider}/{dataset}")
d <- read_csv_files(
provider = provider,
dataset = dataset,
subdir = subdir,
dir_data = dir_data,
sync_archive = TRUE,
metadata_dir = here("metadata"),
field_descriptions = list(
"194903-202105_Cast" = glue(
"{dir_dataset}/Cast Field Descriptions.csv"),
"194903-202105_Bottle" = glue(
"{dir_dataset}/Bottle Field Descriptions - UTF-8.csv")))
# show source files summary
message(glue("Loaded {nrow(d$source_files)} tables from {d$paths$dir_csv}"))
message(glue("Total rows: {sum(d$source_files$nrow)}"))
```
## Check for any mismatched tables and fields
```{r}
#| label: data_integrity_check
# check data integrity
# type_exceptions = "all": type mismatches (34) are expected since readr
# infers types differently (e.g. quality codes as numeric, Time as hms);
# these are resolved during ingestion via flds_redefine.csv type_new
integrity <- check_data_integrity(
d = d,
dataset_name = dataset_name,
type_exceptions = "all")
```
```{r}
#| label: data_integrity_message
#| output: asis
#| echo: false
# render the pass/fail message as markdown
render_integrity_message(integrity)
```
## Show Source Files
```{r}
#| label: source-files
show_source_files(d)
```
## Show CSV Tables and Fields to Ingest
```{r}
#| label: tbls_in
d$d_csv$tables |>
datatable(caption = "Tables to ingest.")
```
```{r}
#| label: flds_in
d$d_csv$fields |>
datatable(caption = "Fields to ingest.")
```
## Show tables and fields redefined
```{r}
#| label: tbls_rd
show_tables_redefine(d)
```
```{r}
#| label: flds_rd
show_fields_redefine(d)
```
## Load Tables into Database
```{r}
#| label: load_tbls_to_db
# use ingest_dataset() which handles:
# - transform_data() for applying redefinitions
# - provenance tracking via gcs_path from read_csv_files()
# - ingest_to_working() for each table
tbl_stats <- ingest_dataset(
con = con,
d = d,
mode = if (overwrite) "replace" else "append",
verbose = TRUE)
tbl_stats |>
datatable(rownames = FALSE, filter = "top")
```
## Apply Ship Corrections
Apply known ship name/code corrections from `ship_renames.csv`.
```{r}
#| label: apply_ship_corrections
ship_renames_csv <- here(
"metadata/calcofi.org/bottle-database/ship_renames.csv")
d_ship_renames <- read_csv(ship_renames_csv)
# apply each correction
n_updated_total <- 0
for (i in seq_len(nrow(d_ship_renames))) {
old_name <- d_ship_renames$csv_name[i]
old_code <- d_ship_renames$csv_code[i]
new_name <- d_ship_renames$csv_name_new[i]
new_code <- d_ship_renames$csv_code_new[i]
# update where both name and code match old values
n_updated <- dbExecute(con, glue_sql(
"UPDATE casts
SET ship_name = {new_name}, ship_code = {new_code}
WHERE ship_name = {old_name} AND ship_code = {old_code}",
.con = con))
if (n_updated > 0) {
message(glue(
"Updated {n_updated} casts: ",
"'{old_name}' [{old_code}] -> '{new_name}' [{new_code}]"))
}
n_updated_total <- n_updated_total + n_updated
}
message(glue("\nTotal casts updated: {n_updated_total}"))
```
## Column Naming Rationale
The column renames in `flds_redefine.csv` follow these db conventions:
- **`*_id`** for integer primary/foreign keys (e.g. `Cst_Cnt` → `cast_id`, `Btl_Cnt` → `bottle_id`)
- **`*_key`** for varchar natural/composite keys (e.g. `Cast_ID` → `cast_key`, `Sta_ID` → `sta_key`)
- Note: `Cruise_ID` → `cruise_key_0` (interim `_0` suffix avoids collision with SWFSC `cruise_key` YYMMKK; discarded after merge)
- **unit suffixes** for measurements (e.g. `Depthm` → `depth_m`)
### Ambiguous column pairs
| Column pair | Why different |
|------------|---------------|
| `Cst_Cnt` (→ `cast_id`) vs `Cast_ID` (→ `cast_key`) | integer counter (1, 2, 3...) vs varchar composite (19-4903CR-HY-060-0930-05400560) |
| `Sta_ID` (→ `sta_key`) vs `DbSta_ID` (→ `db_sta_key`) | "054.0 056.0" (space-separated) vs "05400560" (compact, derivable) |
| `Depth_ID` (→ `depth_key`) vs `Depthm` (→ `depth_m`) | varchar identifier vs numeric depth in meters |
### `R_*` = "Reported" (pre-QC) values
The `R_*` prefix means original instrument values before quality control.
**Critical**: `R_Sal` (Specific Volume Anomaly, values ~233, units 10⁻⁸ m³/kg)
is a **completely different parameter/scale** than `Salnty` (Practical Salinity
PSS-78, values ~33.44). These are pivoted as distinct measurement types
(`r_salinity_sva` vs `salinity`).
## Consolidate Casts Columns
Combine `date` + `time` into `datetime_utc`, drop 16 derivable columns,
and create a `casts_derived` convenience VIEW.
```{r}
#| label: consolidate_casts
# -- create datetime_utc from date (DATE) + time (INTERVAL) ----
# date is DATE type (e.g. 1949-03-01), time is INTERVAL (e.g. 09:30:00)
# adding DATE + INTERVAL yields TIMESTAMP; 324 casts have NULL time
dbExecute(con, "ALTER TABLE casts ADD COLUMN datetime_utc TIMESTAMP")
dbExecute(con, "
UPDATE casts SET datetime_utc =
CASE
WHEN \"time\" IS NOT NULL
THEN CAST(\"date\" AS TIMESTAMP) + \"time\"
ELSE CAST(\"date\" AS TIMESTAMP)
END")
message(glue(
"Created datetime_utc: {dbGetQuery(con, 'SELECT COUNT(*) FROM casts WHERE datetime_utc IS NOT NULL')[[1]]} non-null values"))
# -- drop derivable columns ----
cols_to_drop <- c(
"year", "month", "quarter", "julian_day", "julian_date",
"time_zone",
"lat_deg", "lat_min", "lat_hem",
"lon_deg", "lon_min", "lon_hem",
"cruise", "cruz_sta", "db_sta_key", "cruz_num")
# drop date and time (replaced by datetime_utc)
cols_to_drop <- c(cols_to_drop, "date", "time")
n_before <- length(dbListFields(con, "casts"))
for (col in cols_to_drop) {
tryCatch(
dbExecute(con, glue('ALTER TABLE casts DROP COLUMN "{col}"')),
error = function(e) warning(glue("could not drop {col}: {e$message}")))
}
n_after <- length(dbListFields(con, "casts"))
message(glue("casts columns: {n_before} -> {n_after} (dropped {n_before - n_after})"))
```
## Pivot Bottle Measurements
Pivot wide-format measurement columns from `bottle` into long-format
`bottle_measurement` table. Each measurement type has a value, optional
precision, and optional quality code.
```{mermaid}
%%| label: pivot_bottle_meas_diagram
%%| fig-cap: "Pivot wide bottle columns into long-format bottle_measurement table."
erDiagram
bottle_BEFORE {
int bottle_id PK
int cast_id FK
dbl depth_m
dbl t_deg_c
dbl t_prec
str t_qual
dbl salnty
dbl s_prec
str s_qual
dbl o2ml_l
str o_qual
dbl chlor_a
str chlqua
dbl _etc "... 30 measurement columns"
}
bottle_AFTER {
int bottle_id PK
int cast_id FK
dbl depth_m
str depth_qual
}
bottle_measurement {
int bottle_measurement_id PK
int bottle_id FK
str measurement_type
dbl measurement_value
dbl measurement_prec
str measurement_qual
}
bottle_BEFORE ||--|{ bottle_measurement : "pivot into"
bottle_AFTER ||--o{ bottle_measurement : "bottle_id"
```
```{r}
#| label: pivot_bottle_measurements
# define measurement mapping: type -> value/prec/qual columns
bottle_meas_map <- tribble(
~measurement_type, ~value_col, ~prec_col, ~qual_col,
"temperature", "t_deg_c", "t_prec", "t_qual",
"salinity", "salnty", "s_prec", "s_qual",
"oxygen_ml_l", "o2ml_l", NA, "o_qual",
"sigma_theta", "s_theta", NA, "s_thtaq",
"oxygen_saturation", "o2sat", NA, "o2satq",
"oxygen_umol_kg", "oxy_umol_kg", NA, NA,
"chlorophyll_a", "chlor_a", NA, "chlqua",
"phaeopigment", "phaeop", NA, "phaqua",
"phosphate", "po4u_m", NA, "po4q",
"silicate", "si_o3u_m", NA, "si_o3qu",
"nitrite", "no2u_m", NA, "no2q",
"nitrate", "no3u_m", NA, "no3q",
"ammonia", "nh3u_m", NA, "nh3q",
"c14_rep1", "c14as1", "c14a1p", "c14a1q",
"c14_rep2", "c14as2", "c14a2p", "c14a2q",
"c14_dark", "dark_as", "dark_ap", "darkaq",
"c14_mean", "mean_as", "mean_ap", "mean_aq",
"light_pct", "light_p", NA, NA,
"r_depth", "r_depth", NA, NA,
"r_temperature", "r_temp", NA, NA,
"r_salinity_sva", "r_sal", NA, NA,
"r_dynamic_height", "r_dynht", NA, NA,
"r_ammonium", "r_nuts", NA, NA,
"r_oxygen_umol_kg", "r_oxy_umol_kg", NA, NA,
"dic_rep1", "dic1", NA, "dic_quality_comment",
"dic_rep2", "dic2", NA, "dic_quality_comment",
"alkalinity_rep1", "ta1", NA, NA,
"alkalinity_rep2", "ta2", NA, NA,
"ph_rep1", "p_h1", NA, NA,
"ph_rep2", "p_h2", NA, NA)
# build UNION ALL SQL from mapping
sql_parts <- purrr::pmap_chr(bottle_meas_map, function(
measurement_type, value_col, prec_col, qual_col) {
prec_expr <- if (is.na(prec_col)) "NULL" else prec_col
qual_expr <- if (is.na(qual_col)) "NULL" else qual_col
glue(
"SELECT bottle_id, '{measurement_type}' AS measurement_type,
CAST({value_col} AS DOUBLE) AS measurement_value,
CAST({prec_expr} AS DOUBLE) AS measurement_prec,
CAST({qual_expr} AS VARCHAR) AS measurement_qual
FROM bottle WHERE {value_col} IS NOT NULL")
})
sql_create <- glue(
"CREATE TABLE bottle_measurement AS
SELECT ROW_NUMBER() OVER (ORDER BY bottle_id, measurement_type)
AS bottle_measurement_id, *
FROM (
{paste(sql_parts, collapse = '\nUNION ALL\n')}
) sub")
dbExecute(con, sql_create)
n_meas <- dbGetQuery(con, "SELECT COUNT(*) FROM bottle_measurement")[[1]]
n_types <- dbGetQuery(con, "SELECT COUNT(DISTINCT measurement_type) FROM bottle_measurement")[[1]]
message(glue("bottle_measurement: {format(n_meas, big.mark = ',')} rows, {n_types} measurement types"))
# -- drop measurement columns from bottle base table ----
cols_to_drop <- unique(c(
bottle_meas_map$value_col,
na.omit(bottle_meas_map$prec_col),
na.omit(bottle_meas_map$qual_col)))
n_before <- length(dbListFields(con, "bottle"))
for (col in cols_to_drop) {
tryCatch(
dbExecute(con, glue('ALTER TABLE bottle DROP COLUMN "{col}"')),
error = function(e) warning(glue("could not drop {col}: {e$message}")))
}
# rename p_qual to depth_qual (pressure/depth quality code stays in base table)
dbExecute(con, 'ALTER TABLE bottle RENAME COLUMN p_qual TO depth_qual')
n_after <- length(dbListFields(con, "bottle"))
message(glue("bottle columns: {n_before} -> {n_after} (dropped {n_before - n_after})"))
```
## Pivot Cast Conditions
Pivot meteorological and environmental observation columns from `casts`
into long-format `cast_condition` table.
```{mermaid}
%%| label: pivot_cast_cond_diagram
%%| fig-cap: "Pivot wide cast condition columns into long-format cast_condition table."
erDiagram
casts_BEFORE {
int cast_id PK
str sta_key
dbl lat_dec
dbl lon_dec
ts datetime_utc
dbl wave_dir
dbl wave_ht
dbl wave_prd
dbl wind_dir
dbl wind_spd
dbl barometer
dbl dry_t
dbl wet_t
dbl wea
dbl cloud_typ
dbl cloud_amt
dbl visibility
dbl secchi
dbl forel_u
}
casts_AFTER {
int cast_id PK
str sta_key
dbl lat_dec
dbl lon_dec
ts datetime_utc
}
cast_condition {
int cast_condition_id PK
int cast_id FK
str condition_type
dbl condition_value
}
casts_BEFORE ||--|{ cast_condition : "pivot into"
casts_AFTER ||--o{ cast_condition : "cast_id"
```
```{r}
#| label: pivot_cast_conditions
cast_cond_map <- tribble(
~condition_type, ~source_col,
"wave_direction", "wave_dir",
"wave_height", "wave_ht",
"wave_period", "wave_prd",
"wind_direction", "wind_dir",
"wind_speed", "wind_spd",
"barometric_pressure", "barometer",
"dry_air_temp", "dry_t",
"wet_air_temp", "wet_t",
"weather_code", "wea",
"cloud_type", "cloud_typ",
"cloud_amount", "cloud_amt",
"visibility", "visibility",
"secchi_depth", "secchi",
"water_color", "forel_u")
# build UNION ALL SQL
sql_parts <- purrr::pmap_chr(cast_cond_map, function(condition_type, source_col) {
glue(
"SELECT cast_id, '{condition_type}' AS condition_type,
CAST({source_col} AS DOUBLE) AS condition_value
FROM casts WHERE {source_col} IS NOT NULL")
})
sql_create <- glue(
"CREATE TABLE cast_condition AS
SELECT ROW_NUMBER() OVER (ORDER BY cast_id, condition_type)
AS cast_condition_id, *
FROM (
{paste(sql_parts, collapse = '\nUNION ALL\n')}
) sub")
dbExecute(con, sql_create)
n_cond <- dbGetQuery(con, "SELECT COUNT(*) FROM cast_condition")[[1]]
n_types <- dbGetQuery(con, "SELECT COUNT(DISTINCT condition_type) FROM cast_condition")[[1]]
message(glue("cast_condition: {format(n_cond, big.mark = ',')} rows, {n_types} condition types"))
# -- drop condition columns from casts ----
n_before <- length(dbListFields(con, "casts"))
for (col in cast_cond_map$source_col) {
tryCatch(
dbExecute(con, glue('ALTER TABLE casts DROP COLUMN "{col}"')),
error = function(e) warning(glue("could not drop {col}: {e$message}")))
}
n_after <- length(dbListFields(con, "casts"))
message(glue("casts columns: {n_before} -> {n_after} (dropped {n_before - n_after})"))
```
## Load Measurement Type Reference
Load the `measurement_type` lookup table that provides units and
descriptions for all measurement/condition types across datasets.
### `measurement_type` vs `lookup`
The `measurement_type` table and the swfsc `lookup` table serve fundamentally different purposes:
- **`measurement_type`** answers "what was measured?" — provides metadata (units, description, source provenance) for measurement codes used in `bottle_measurement`, `cast_condition`, and `ichthyo`
- **`lookup`** answers "what does this code mean?" — translates categorical values (egg developmental stages 1-11, larva stages, tow types) into human-readable descriptions
Merging would create a sparse table with many NULL columns (e.g., `units`/`_source_column` NULL for egg stages; `lookup_num`/`lookup_chr` NULL for temperature). Both tables coexist in the Working DuckLake, loaded from their respective upstream workflows.
```{r}
#| label: load_measurement_type
meas_type_csv <- here(
"metadata/calcofi.org/bottle-database/measurement_type.csv")
d_meas_type <- read_csv(meas_type_csv)
dbWriteTable(con, "measurement_type", d_meas_type, overwrite = TRUE)
# validate FK: all bottle_measurement types exist in reference table
orphan_btl_types <- dbGetQuery(con, "
SELECT DISTINCT measurement_type FROM bottle_measurement
WHERE measurement_type NOT IN (SELECT measurement_type FROM measurement_type)")
stopifnot(
"all bottle_measurement.measurement_type must exist in measurement_type" =
nrow(orphan_btl_types) == 0)
# validate FK: all cast_condition types exist in reference table
orphan_cond_types <- dbGetQuery(con, "
SELECT DISTINCT condition_type FROM cast_condition
WHERE condition_type NOT IN (SELECT measurement_type FROM measurement_type)")
stopifnot(
"all cast_condition.condition_type must exist in measurement_type" =
nrow(orphan_cond_types) == 0)
message(glue("measurement_type: {nrow(d_meas_type)} types loaded, FK checks passed"))
```
## Verify Primary Keys
```{r}
#| label: verify_primary_keys
# verify cast_id uniqueness in casts
cast_keys <- tbl(con, "casts") |> pull(cast_id)
n_dup_cast <- sum(duplicated(cast_keys))
stopifnot("cast_id must be unique in casts" = n_dup_cast == 0)
message(glue("casts.cast_id: {length(cast_keys)} unique values, 0 duplicates"))
# verify bottle_id uniqueness in bottle
btl_keys <- tbl(con, "bottle") |> pull(bottle_id)
n_dup_btl <- sum(duplicated(btl_keys))
stopifnot("bottle_id must be unique in bottle" = n_dup_btl == 0)
message(glue("bottle.bottle_id: {length(btl_keys)} unique values, 0 duplicates"))
# verify FK: bottle.cast_id -> casts.cast_id
orphan_bottles <- tbl(con, "bottle") |>
anti_join(
tbl(con, "casts"),
by = "cast_id") |>
collect()
stopifnot(
"all bottle.cast_id must exist in casts.cast_id" =
nrow(orphan_bottles) == 0)
message(glue(
"FK integrity: all {length(btl_keys)} bottle.cast_id ",
"found in casts.cast_id"))
# verify bottle_measurement_id uniqueness
n_dup_bm_id <- dbGetQuery(con, "
SELECT COUNT(*) FROM (
SELECT bottle_measurement_id, COUNT(*) AS n
FROM bottle_measurement
GROUP BY bottle_measurement_id
HAVING COUNT(*) > 1)")[[1]]
stopifnot("bottle_measurement_id must be unique" = n_dup_bm_id == 0)
message(glue("bottle_measurement.bottle_measurement_id: unique, 0 duplicates"))
# verify composite PK: bottle_measurement (bottle_id, measurement_type)
n_dup_meas <- dbGetQuery(con, "
SELECT COUNT(*) FROM (
SELECT bottle_id, measurement_type, COUNT(*) AS n
FROM bottle_measurement
GROUP BY bottle_id, measurement_type
HAVING COUNT(*) > 1)")[[1]]
stopifnot("bottle_measurement (bottle_id, measurement_type) must be unique" = n_dup_meas == 0)
message(glue("bottle_measurement: composite key unique, 0 duplicates"))
# verify FK: bottle_measurement.bottle_id -> bottle.bottle_id
n_orphan_meas <- dbGetQuery(con, "
SELECT COUNT(DISTINCT bottle_id) FROM bottle_measurement
WHERE bottle_id NOT IN (SELECT bottle_id FROM bottle)")[[1]]
stopifnot("all bottle_measurement.bottle_id must exist in bottle" = n_orphan_meas == 0)
message(glue("bottle_measurement FK: all bottle_ids found in bottle"))
# verify cast_condition_id uniqueness
n_dup_cc_id <- dbGetQuery(con, "
SELECT COUNT(*) FROM (
SELECT cast_condition_id, COUNT(*) AS n
FROM cast_condition
GROUP BY cast_condition_id
HAVING COUNT(*) > 1)")[[1]]
stopifnot("cast_condition_id must be unique" = n_dup_cc_id == 0)
message(glue("cast_condition.cast_condition_id: unique, 0 duplicates"))
# verify composite PK: cast_condition (cast_id, condition_type)
n_dup_cond <- dbGetQuery(con, "
SELECT COUNT(*) FROM (
SELECT cast_id, condition_type, COUNT(*) AS n
FROM cast_condition
GROUP BY cast_id, condition_type
HAVING COUNT(*) > 1)")[[1]]
stopifnot("cast_condition (cast_id, condition_type) must be unique" = n_dup_cond == 0)
message(glue("cast_condition: composite key unique, 0 duplicates"))
# verify FK: cast_condition.cast_id -> casts.cast_id
n_orphan_cond <- dbGetQuery(con, "
SELECT COUNT(DISTINCT cast_id) FROM cast_condition
WHERE cast_id NOT IN (SELECT cast_id FROM casts)")[[1]]
stopifnot("all cast_condition.cast_id must exist in casts" = n_orphan_cond == 0)
message(glue("cast_condition FK: all cast_ids found in casts"))
```
## Schema Documentation
```{r}
#| label: dm_tbls
# exclude views (casts_derived) — dm_from_con can't access views
db_tables <- function(con) {
setdiff(dbListTables(con), "casts_derived") |> sort()
}
tbls <- db_tables(con)
# dm object for visualization (keys learned from data, not DB constraints)
dm_dev <- dm_from_con(con, table_names = tbls, learn_keys = FALSE)
dm_draw(dm_dev, view_type = "all")
```
### Primary Key Strategy
**Note on UUID-first approach**: The SWFSC ichthyo tables use source UUIDs (minted
at sea) as primary identifiers. The bottle/cast tables are different — they use
source integer counters (`Cst_Cnt`, `Btl_Cnt`) as natural keys, which are stable
identifiers from the source system. The pivoted tables (`bottle_measurement`,
`cast_condition`) have no source identifier and use sequential integer IDs for
convenience, with composite natural keys enforced as unique.
| Table | Primary Key | Type |
|-------|-------------|------|
| `casts` | `cast_id` | Source integer counter (`Cst_Cnt`) — stable source key |
| `bottle` | `bottle_id` | Source integer counter (`Btl_Cnt`) — stable source key |
| `bottle_measurement` | `bottle_measurement_id` | Sequential (derived/pivoted table); composite `(bottle_id, measurement_type)` enforced unique |
| `cast_condition` | `cast_condition_id` | Sequential (derived/pivoted table); composite `(cast_id, condition_type)` enforced unique |
| `measurement_type` | `measurement_type` | Natural key (type code string) |
| `grid` | `grid_key` | Natural key |
### Foreign Key Relationships
```
casts.cast_id (PK, integer) — station-level
↓
bottle.bottle_id (PK, integer) — sample-level
bottle.cast_id (FK) → casts.cast_id
↓
bottle_measurement.bottle_measurement_id (PK, integer) — ROW_NUMBER(ORDER BY bottle_id, measurement_type)
bottle_measurement.bottle_id (FK) → bottle.bottle_id
bottle_measurement.measurement_type (FK) → measurement_type.measurement_type
cast_condition.cast_condition_id (PK, integer) — ROW_NUMBER(ORDER BY cast_id, condition_type)
cast_condition.cast_id (FK) → casts.cast_id
cast_condition.condition_type (FK) → measurement_type.measurement_type
casts.grid_key (FK) → grid.grid_key (after spatial join)
casts.ship_code (soft FK) → ship.ship_nodc (validated after integration with swfsc)
```
```{r}
#| label: dm_fk
tbls <- db_tables(con)
dm_dev <- dm_from_con(con, table_names = tbls, learn_keys = FALSE)
# define relationships in dm (in-memory for visualization only)
add_bottle_keys <- function(dm){
dm |>
dm_add_pk(casts, cast_id) |>
dm_add_pk(bottle, bottle_id) |>
dm_add_pk(bottle_measurement, bottle_measurement_id) |>
dm_add_pk(cast_condition, cast_condition_id) |>
dm_add_pk(measurement_type, measurement_type) |>
dm_add_fk(bottle, cast_id, casts) |>
dm_add_fk(bottle_measurement, bottle_id, bottle) |>
dm_add_fk(bottle_measurement, measurement_type, measurement_type) |>
dm_add_fk(cast_condition, cast_id, casts) |>
dm_add_fk(cast_condition, condition_type, measurement_type, measurement_type)
}
dm_dev_fk <- dm_dev |>
add_bottle_keys()
dm_draw(dm_dev_fk, view_type = "all")
```
## Add Spatial
### Add `casts.geom`
```{r}
#| label: mk_cast_pts
# add geometry column using DuckDB spatial
# note: DuckDB spatial doesn't track SRID metadata (unlike PostGIS)
# all geometries assumed WGS84 (EPSG:4326) by convention
add_point_geom(con, "casts")
```
### Load `grid` table
Load grid from the swfsc workflow's GCS parquet output (single source of
truth — grid is created in `ingest_swfsc.noaa.gov_calcofi-db.qmd`).
```{r}
#| label: load_grid
load_gcs_parquet_to_duckdb(
con = con,
gcs_path = "gs://calcofi-db/ingest/swfsc.noaa.gov_calcofi-db/grid.parquet",
table_name = "grid")
```
### Update `casts.grid_key`
```{r}
#| label: update_cast_from_grid
grid_stats <- assign_grid_key(con, "casts")
grid_stats |> datatable()
```
### Create `casts_derived` VIEW
Create convenience VIEW after all casts table modifications are complete
(column drops, pivots, spatial join). The VIEW adds back temporal and
coordinate columns derivable from the base table.
```{r}
#| label: create_casts_derived
dbExecute(con, "DROP VIEW IF EXISTS casts_derived")
dbExecute(con, "
CREATE VIEW casts_derived AS
SELECT *,
EXTRACT(YEAR FROM datetime_utc)::SMALLINT AS year,
EXTRACT(MONTH FROM datetime_utc)::SMALLINT AS month,
EXTRACT(QUARTER FROM datetime_utc)::SMALLINT AS quarter,
EXTRACT(DOY FROM datetime_utc)::SMALLINT AS julian_day,
(datetime_utc::DATE - DATE '1899-12-30') AS julian_date,
FLOOR(ABS(lat_dec))::SMALLINT AS lat_deg,
(ABS(lat_dec) - FLOOR(ABS(lat_dec))) * 60 AS lat_min,
CASE WHEN lat_dec >= 0 THEN 'N' ELSE 'S' END AS lat_hem,
FLOOR(ABS(lon_dec))::SMALLINT AS lon_deg,
(ABS(lon_dec) - FLOOR(ABS(lon_dec))) * 60 AS lon_min,
CASE WHEN lon_dec >= 0 THEN 'E' ELSE 'W' END AS lon_hem,
STRFTIME(datetime_utc, '%Y%m') AS cruise,
REPLACE(REPLACE(sta_key, '.', ''), ' ', '') AS db_sta_key
FROM casts")
message("Created casts_derived VIEW with derived temporal/coordinate columns")
```
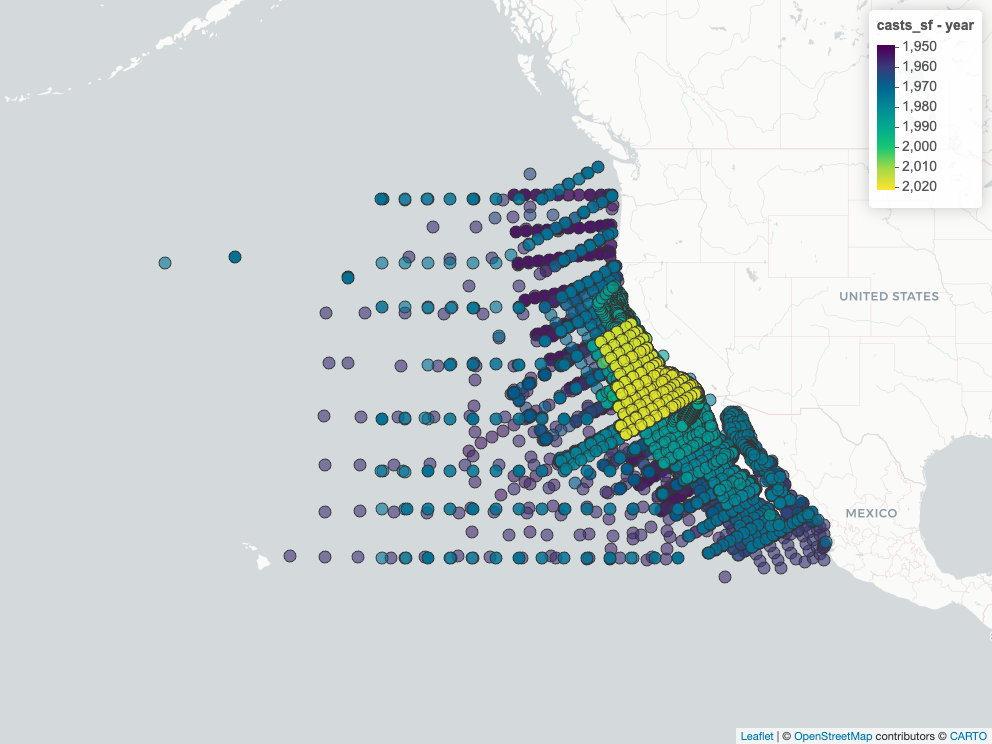
### Map Cast Locations
```{r}
#| label: map_casts
# slowish, so use cached figure
map_casts_png <- here(glue("figures/{provider}_{dataset}_casts_map.png"))
if (!file_exists(map_casts_png)) {
casts_sf <- calcofi4r::cc_read_sf(con, "casts") |>
mutate(year = year(datetime_utc))
m <- mapView(casts_sf, zcol = "year")
mapshot2(m, file = map_casts_png)
}
htmltools::img(
src = map_casts_png |> str_replace(here(), "."),
width = "600px")
```
## Report
```{r}
#| label: show_latest
all_tbls <- db_tables(con)
add_bottle_spatial_keys <- function(dm){
dm |>
dm_add_pk(grid, grid_key) |>
dm_add_fk(casts, grid_key, grid)
}
dm_final <- dm_from_con(con, table_names = all_tbls, learn_keys = FALSE) |>
add_bottle_keys() |>
add_bottle_spatial_keys() |>
dm_set_colors(
lightgreen = grid,
lightyellow = c(bottle_measurement, cast_condition),
lightblue = measurement_type)
dm_draw(dm_final, view_type = "all")
```
```{r}
#| label: summary_stats
# summary statistics
n_casts <- tbl(con, "casts") |> tally() |> pull(n)
n_bottles <- tbl(con, "bottle") |> tally() |> pull(n)
n_meas <- dbGetQuery(con, "SELECT COUNT(*) FROM bottle_measurement")[[1]]
n_cond <- dbGetQuery(con, "SELECT COUNT(*) FROM cast_condition")[[1]]
year_range <- dbGetQuery(con, "
SELECT
EXTRACT(YEAR FROM MIN(datetime_utc))::INTEGER AS yr_min,
EXTRACT(YEAR FROM MAX(datetime_utc))::INTEGER AS yr_max
FROM casts")
fmt <- function(x, ...) format(x, big.mark = ",", ...)
message(glue("Total casts: {fmt(n_casts)}"))
message(glue("Total bottles: {fmt(n_bottles)}"))
message(glue("Total bottle measurements: {fmt(n_meas)}"))
message(glue("Total cast conditions: {fmt(n_cond)}"))
message(glue("Year range: {year_range$yr_min} - {year_range$yr_max}"))
```
## Validate Local Database
Validate data quality in the local wrangling database before exporting
to parquet. The parquet outputs from this workflow can later be used to
update the Working DuckLake.
```{r}
#| label: validate
# validate data quality
validation <- validate_for_release(con)
if (validation$passed) {
message("Validation passed!")
if (nrow(validation$checks) > 0) {
validation$checks |>
filter(status != "pass") |>
datatable(caption = "Validation Warnings")
}
} else {
cat("Validation FAILED:\n")
cat(paste("-", validation$errors, collapse = "\n"))
}
```
## Enforce Column Types
Force integer/smallint types on columns that R's `numeric` mapped to
`DOUBLE` during `dbWriteTable()`. Uses `flds_redefine.csv` (`type_new`)
as the source of truth for source-table columns, plus explicit overrides
for derived-table columns.
```{r}
#| label: enforce_types
type_changes <- enforce_column_types(
con = con,
d_flds_rd = d$d_flds_rd,
type_overrides = list(),
tables = db_tables(con),
verbose = TRUE)
if (nrow(type_changes) > 0) {
type_changes |>
datatable(caption = "Column type changes applied")
}
# recreate casts_derived VIEW after type changes
dbExecute(con, "DROP VIEW IF EXISTS casts_derived")
dbExecute(con, "
CREATE VIEW casts_derived AS
SELECT *,
EXTRACT(YEAR FROM datetime_utc)::SMALLINT AS year,
EXTRACT(MONTH FROM datetime_utc)::SMALLINT AS month,
EXTRACT(QUARTER FROM datetime_utc)::SMALLINT AS quarter,
EXTRACT(DOY FROM datetime_utc)::SMALLINT AS julian_day,
(datetime_utc::DATE - DATE '1899-12-30') AS julian_date,
FLOOR(ABS(lat_dec))::SMALLINT AS lat_deg,
(ABS(lat_dec) - FLOOR(ABS(lat_dec))) * 60 AS lat_min,
CASE WHEN lat_dec >= 0 THEN 'N' ELSE 'S' END AS lat_hem,
FLOOR(ABS(lon_dec))::SMALLINT AS lon_deg,
(ABS(lon_dec) - FLOOR(ABS(lon_dec))) * 60 AS lon_min,
CASE WHEN lon_dec >= 0 THEN 'E' ELSE 'W' END AS lon_hem,
STRFTIME(datetime_utc, '%Y%m') AS cruise,
REPLACE(REPLACE(sta_key, '.', ''), ' ', '') AS db_sta_key
FROM casts")
message("Recreated casts_derived VIEW after column type enforcement")
```
## Data Preview
Preview first and last rows of each table before writing parquet outputs.
```{r}
#| label: preview_tables
#| results: asis
preview_tables(con, c(
"casts", "bottle", "bottle_measurement",
"cast_condition", "measurement_type")) # grid canonical from swfsc
```
```{r}
#| label: preview_casts_derived
# casts_derived is a VIEW, preview separately
dbGetQuery(con, "SELECT * FROM casts_derived LIMIT 100") |>
datatable(
caption = "casts_derived — first 100 rows (VIEW)",
rownames = FALSE,
filter = "top")
```
## Write Parquet Outputs
Export tables to parquet files for downstream use.
```{r}
#| label: write_parquet
# write parquet files with manifest
parquet_stats <- write_parquet_outputs(
con = con,
output_dir = dir_parquet,
tables = setdiff(db_tables(con), "grid"), # grid canonical from swfsc
strip_provenance = F)
parquet_stats |>
mutate(file = basename(path)) |>
select(-path) |>
datatable(caption = "Parquet export statistics")
```
## Write Metadata
Build `metadata.json` sidecar file documenting all tables and columns in
parquet outputs. DuckDB `COMMENT ON` does not propagate to parquet, so
this provides the metadata externally.
```{r}
#| label: write_metadata
metadata_path <- build_metadata_json(
con = con,
d_tbls_rd = d$d_tbls_rd,
d_flds_rd = d$d_flds_rd,
metadata_derived_csv = here(
"metadata/calcofi.org/bottle-database/metadata_derived.csv"),
output_dir = dir_parquet,
tables = db_tables(con),
set_comments = TRUE,
provider = provider,
dataset = dataset,
workflow_url = glue(
"https://calcofi.io/workflows/",
"ingest_calcofi.org_bottle-database.html"))
# show metadata summary
metadata <- jsonlite::fromJSON(metadata_path)
tibble(
table = names(metadata$tables),
n_cols = map_int(
names(metadata$tables),
~sum(grepl(glue("^{.x}\\."), names(metadata$columns)))),
name_long = map_chr(metadata$tables, ~.x$name_long)) |>
datatable(caption = "Table metadata summary")
```
```{r}
#| label: show_metadata_json
listviewer::jsonedit(
jsonlite::fromJSON(metadata_path, simplifyVector = FALSE),
mode = "view")
```
## Upload to GCS Archive
Upload parquet files, manifest, and metadata sidecar to
`gs://calcofi-db/ingest/{provider}_{dataset}/`.
```{r}
#| label: upload_gcs
gcs_ingest_prefix <- glue("ingest/{provider}_{dataset}")
gcs_bucket <- "calcofi-db"
# list local files to upload (parquet + json sidecars)
local_files <- list.files(dir_parquet, full.names = TRUE)
# upload each file
walk(local_files, function(f) {
gcs_path <- glue("gs://{gcs_bucket}/{gcs_ingest_prefix}/{basename(f)}")
put_gcs_file(f, gcs_path)
message(glue("Uploaded {basename(f)} -> {gcs_path}"))
})
message(glue(
"\nUploaded {length(local_files)} files to ",
"gs://{gcs_bucket}/{gcs_ingest_prefix}/"))
```
## Cleanup
```{r}
#| label: cleanup
# close local wrangling database connection
close_duckdb(con)
message("Local wrangling database connection closed")
# note: parquet outputs are in data/parquet/calcofi.org_bottle-database/
# these can be used to update the Working DuckLake in a separate workflow
message(glue("Parquet outputs written to: {dir_parquet}"))
message(glue("GCS outputs at: gs://{gcs_bucket}/{gcs_ingest_prefix}/"))
```
## TODO
- [x] Review column names and apply db naming conventions, per @docs/db.qmd and @workflows/README_PLAN.qmd: cast_cnt -> cast_id, btl_cnt -> bottle_id, sta_id -> sta_key, etc.
- [x] Pivot longer casts/bottle tables for easier analysis. Created bottle_measurement (value/prec/qual) and cast_condition tables, plus measurement_type reference.
- [x] Consolidate casts temporal columns (date+time -> datetime_utc), drop 16 derivable columns, create casts_derived VIEW.
- [ ] Link possible fields to @workflows/ingest_swfsc.noaa.gov_calcofi-db.qmd. Flag mismatches.
- [x] Cruise bridge prep: renamed `Cruise_ID` → `cruise_key_0` (was `cruise_key`) to avoid collision with SWFSC `cruise_key` (YYMMKK). Merge workflow adds `cruise_key` as new column derived from `datetime_utc` + `ship_key`.
- [ ] Validate casts.ship_code -> ship.ship_nodc FK after integration with swfsc data.
- [ ] Review documentation for more comment descriptions at [Data \>
Data Formats \| CalCOFI.org](https://calcofi.com/index.php?option=com_content&view=category&id=73&Itemid=993)
::: {.callout-caution collapse="true"}
## Session Info
```{r session_info}
devtools::session_info()
```
:::