---
title: "Interpolate Larvae"
author: "Ben Best"
format:
html:
toc: true
toc-depth: 3
code-fold: true
editor_options:
chunk_output_type: console
---
## CalCOFI Larvae
Constraints described by Andrew Thompson in CalCOFI Analysis for CINMS/MBNMS infographics ([calcofi.io/larvae-cinms](https://calcofi.io/larvae-cinms)):
- select only starboard samples from the bongo nets\
`net2cruise.netside = 'S'` (Port is usually ethanol sample vs formalin)
- select a minimum date\
`net2cruise.cruise_ymd >= '19780101'` \
diff't gear type, like oblique tow. internal table for
Questions:
- Should we limit (or have option) to filter by `linestation_core`?
- Should we limit (or have option) to limit by towtype?
```r
tbl(con, "net2cruise") |> pull(towtype) |> table()
```
```
C1 CB CV MT PV
28,640 18,819 6,402 12,757 12,065
```
See `tow_types` table:
- CB: standard bongo tow starting 1978-01-01
- C1: previously used ringnet
### Species Groups
```{r larvae}
librarian::shelf(
calcofi/calcofi4r,
dplyr, DT, dygraphs, glue, here, mapview, purrr, readr, sf, tidyr,
quiet = T)
source(here("../apps_dev/libs/db.R")) # con: database connection
tbl(con, "species_groups") |>
group_by(spp_group) |>
summarize(n_spp = n()) |>
collect() |>
datatable()
```
### Anchovy by Year-Month
```{r larvae-anchovy-ym}
spp_group <- "Anchovy"
ym <- "2005-04"
monthly_csv <- here(glue("data/larvae_{spp_group}-ym.csv"))
ym_geo <- here(glue("data/larvae_{spp_group}-{ym}.geojson"))
if (any(!file.exists(c(monthly_csv, ym_geo)))){
d <- tbl(con, "net2cruise") |> # colnames() |> paste(collapse=", ")
# netid, cruise_id, cruise_ymd, ship, line, stationid, station, latitude, longitude, orderocc, gebco_depth, netside, townumber, towtype, volsampled, shf, propsorted, starttime, geom
filter(
netside == "S",
cruise_ymd >= "19780101") |>
left_join(
tbl(con, "larvae_species"),
# netid, spccode, scientific_name, common_name, larvaecount, itis_tsn
by="netid") |>
left_join(
tbl(con, "species_groups") |>
# spp_group, spccode
filter(spp_group == !!spp_group),
by="spccode") |>
select(
netid, longitude, latitude, starttime, volsampled, # net2cruise
spccode, scientific_name, common_name, larvaecount, itis_tsn, # larvae_species
spp_group) |> # species_groups
arrange(starttime) |>
collect() |> # 27,721 × 6 <- 145,724 × 11
group_by(
netid, longitude, latitude, starttime, volsampled) |>
nest() |>
mutate(
larvaecount = map_int(data, \(x){
x |>
filter(
!is.na(spp_group)) |>
summarize(
n = sum(larvaecount, na.rm = T)) |>
pull(n) } ) )
d_monthly <- d |>
select(-data) |>
mutate(
ym_chr = format(starttime, "%Y-%m"),
ym = glue("{ym_chr}-01") |> as.Date()) |>
group_by(ym) |>
summarize(
vol = first(volsampled),
n = sum(larvaecount)) |>
mutate(
n_log10 = log10(n + 0.001),
n_per_vol = n / vol,
n_per_vol_log10 = log10(n_per_vol + 0.0001))
write_csv(d_monthly, monthly_csv)
pts_ym <- d |> # 99 × 7 <- 27,721 × 7
filter(
format(starttime, "%Y-%m") == "2005-04") |>
select(-data) |>
rename(
n = larvaecount,
vol = volsampled) |>
mutate(
n_log10 = log10(n + 0.001),
n_per_vol = n / vol,
n_per_vol_log10 = log10(n_per_vol + 0.0001)) |>
sf::st_as_sf(
coords = c("longitude", "latitude"),
crs = 4326,
remove = F)
write_sf(pts_ym, ym_geo, delete_dsn = T)
}
d_monthly <- read_csv(monthly_csv, show_col_types = F)
d_monthly |>
select(ym, n_per_vol) |>
arrange(ym) |>
dygraph(
x = "ym",
y = "n_per_vol",
main = glue("Larvae Counts by Month for {spp_group}"))
```
### Map of Anchovy Larvae on 2005-04
```{r anchovy-map-2005-04}
pts_ym <- read_sf(ym_geo)
mapView(
pts_ym,
zcol = "n_per_vol_log10",
col.regions = rev(RColorBrewer::brewer.pal(11,"Spectral")))
```
## Copernicus Marine
- [Global Ocean Physics Reanalysis | Copernicus Marine Service](https://data.marine.copernicus.eu/product/GLOBAL_MULTIYEAR_PHY_001_030/description)
> The GLORYS12V1 product is the CMEMS global ocean eddy-resolving (1/12° horizontal resolution, 50 vertical levels) reanalysis covering the altimetry (1993 onward)...
> This product includes daily and monthly mean files for temperature, salinity, currents, sea level, mixed layer depth and ice parameters from the top to the bottom. The global ocean output files are displayed on a standard regular grid at 1/12° (approximatively 8 km) and on 50 standard levels.
### Visualizing Copernicus in 4D (lon, lat, depth, time)
The Web Map Tile Service (WMTS) allows for interactive map rendering by `variable`, `time` and `depth`. It does not include the raw data values, so is ony for visualization.
The [WMTS `GetCapabilities`](https://wmts.marine.copernicus.eu/teroWmts/GLOBAL_MULTIYEAR_PHY_001_030?request=GetCapabilities) gets parsed to collect available variables, depths (`elevation`) and dates (`time`) for the given `product_id/dataset_id/variable`.
See:
- code: [github.com/CalCOFI/apps:copernicus/](https://github.com/CalCOFI/apps/tree/main/copernicus)
- app: [shiny.calcofi.io/copernicus](https://shiny.calcofi.io/copernicus/)
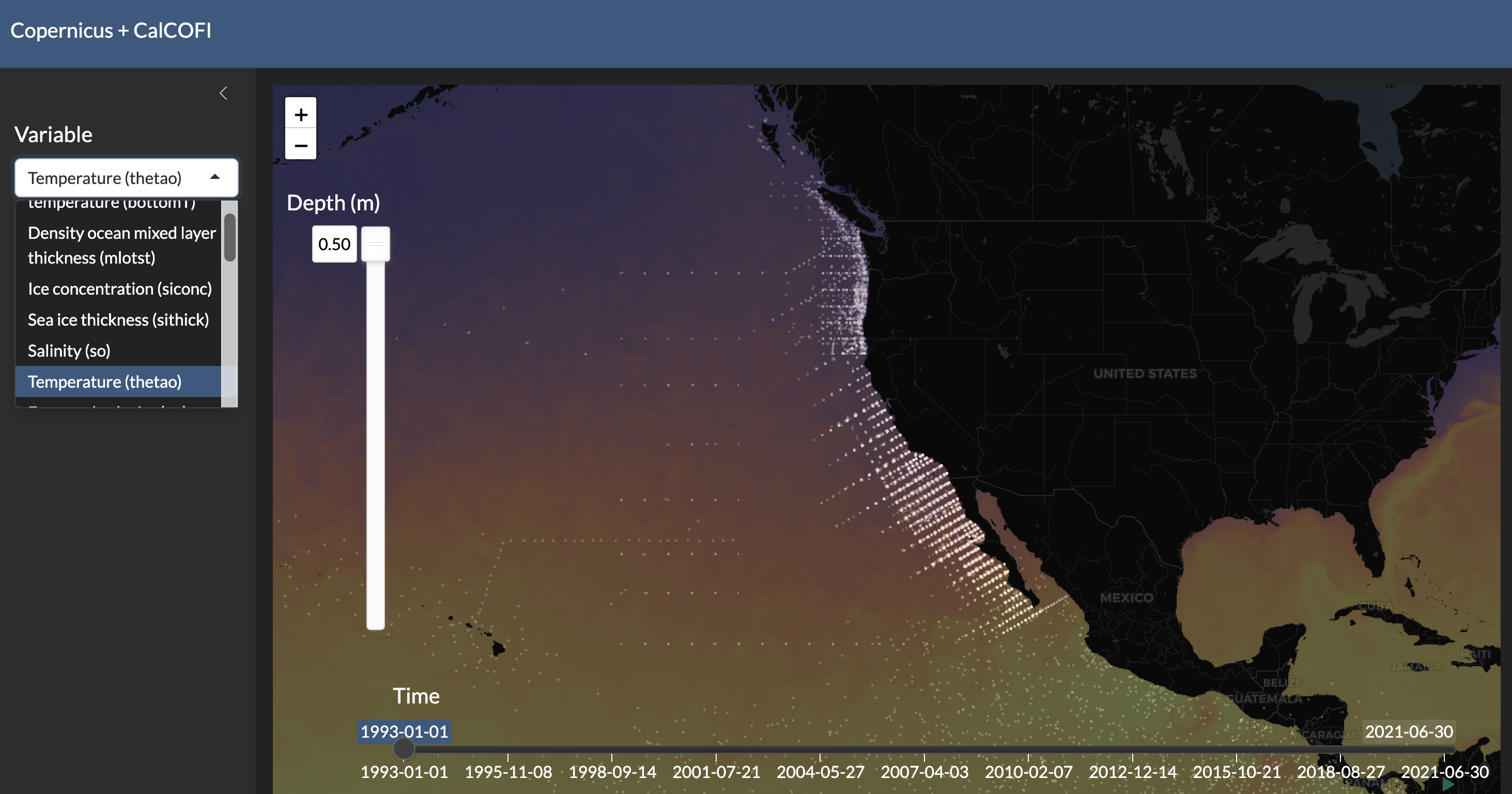
### Subset raster of surface temperature for 2005-04
Using the CLI.
```{r cm-setup}
librarian::shelf(
glue, httr2, leaflet, lubridate, xml2)
# Global Ocean Physics Reanalysis
# 1993-01-01 to 2021-06-01
product_id = "GLOBAL_MULTIYEAR_PHY_001_030"
# dataset_id = "cmems_mod_glo_phy_my_0.083deg_P1D-m" # P1D: daily
dataset_id = "cmems_mod_glo_phy_my_0.083deg_P1M-m" # P1M: monthly
# Global Ocean Physics Analysis and Forecast
# 1 Nov 2020 to 31 May 2024
# product_id = "GLOBAL_ANALYSISFORECAST_PHY_001_024"
# dataset_id = "cmems_mod_glo_phy-thetao_anfc_0.083deg_P1M-m"
# my: MY (Multi-Year/Reprocessed)
# myint: MY interim (MYINT)
# nrt: NRT (Near Real Time)
variable = "thetao"
date = as.Date("2005-04-01") |> as.POSIXct()
```
- `product_id`: `r product_id`
- `dataset_id`: `r dataset_id`
- `variable`: `r variable` (temperature)
```{r cm-cc}
librarian::shelf(
calcofi/calcofi4r,
dplyr, glue, here, jsonlite, leaflet, lubridate, mapview, purrr, sf, terra,
tibble, tidyr)
cm = "/opt/homebrew/Caskroom/mambaforge/base/envs/copernicusmarine/bin/copernicusmarine"
user = "bbest1"
# pass = readLines("~/My Drive/private/data.marine.copernicus.eu_bbest1-password.txt")
pass = "temporary"
verbose = F
# CalCOFI bounding box
b <- st_union(cc_grid_zones) |> st_bbox()
# b
# xmin ymin xmax ymax
# -135.23008 18.42757 -105.77692 49.23891
m_json <- here(glue("data/copernicusmarine/{dataset_id}.json"))
cmd <- glue("{cm} describe --include-datasets -c {dataset_id}")
if (!file.exists(m_json))
system(cmd, intern = TRUE) |> writeLines(m_json)
m_o <- read_json(m_json)
# listviewer::jsonedit(m_o)
get_coord = function(o, v){
if (!v %in% names(o))
return(NA)
if (length(o[[v]]) == 0)
return(NA)
o[[v]] |>
as.character()
}
coord_date <- function(df, col){
df |>
filter(coord_id == "time") |>
pull(!!col) |>
(\(x) as.double(x) / 1000)() |>
as_datetime(origin = "1970-01-01", tz = "UTC") |>
as_date()
}
m_vars <- m_o |>
pluck("products", 1, "datasets", 1, "versions", 1, "parts", 1, "services") |>
keep(\(x) x$service_type$service_name == "arco-geo-series") |>
pluck(1, "variables")
d_vars_coords <- m_vars %>%
tibble(
short_name = map_chr(., "short_name"),
standard_name = map_chr(., "standard_name"),
units = map_chr(., "units"),
coord = map(., "coordinates") ) |>
select(-`.`) |>
unnest(coord) |>
mutate(
coord_id = map_chr(coord, "coordinates_id"),
coord_units = map_chr(coord, "units"),
coord_min_val = map_chr(coord, get_coord, "minimum_value"),
coord_max_val = map_chr(coord, get_coord, "maximum_value"),
coord_step = map_chr(coord, get_coord, "step"),
coord_chunk_length = map_chr(coord, get_coord, "chunking_length"),
coord_chunk_type = map_chr(coord, get_coord, "chunk_type"),
coord_chunk_ref = map_chr(coord, get_coord, "chunk_reference_coordinate"),
coord_chunk_geo = map_chr(coord, get_coord, "chunk_geometric_factor"),
coord_values = map(coord, "values"),
coord_values_n = map_int(coord_values, length))
#
# d_vars_coords |> select(short_name, coord_id) |> table()
# coord_id
# short_name depth latitude longitude time
# bottomT 1 1 1 1
# mlotst 1 1 1 1
# siconc 1 1 1 1
# sithick 1 1 1 1
# so 1 1 1 1
# thetao 1 1 1 1
# uo 1 1 1 1
# usi 1 1 1 1
# vo 1 1 1 1
# vsi 1 1 1 1
# zos 1 1 1 1
#
# d_vars_coords |> select(coord_id, coord_values_n) |> table()
# coord_values_n
# coord_id 0 50
# depth 0 11
# latitude 11 0
# longitude 11 0
# time 11 0
d_vars <- d_vars_coords |>
nest(.by = c(short_name, standard_name, units)) |>
mutate(
date_min = map_dbl(data, coord_date, "coord_min_val") |>
as.Date(),
date_max = map_dbl(data, coord_date, "coord_max_val") |>
as.Date())
depths <- d_vars |>
filter(
short_name == "thetao") |>
pull(data) |>
(\(x) x[[1]])() |>
filter(
coord_id == "depth") |>
pull(coord_values) |>
unlist()
# depths <- obj$products[[1]]$datasets[[1]]$versions[[1]]$parts[[1]]$services[[2]]$variables[[1]]$coordinates[[4]][["values"]] |>
# unlist()
depth <- depths[length(depths)] * -1
# output filename
nc <- here(glue("data/copernicusmarine/{dataset_id}__{variable}__{date}.nc"))
# https://help.marine.copernicus.eu/en/articles/7970514-copernicus-marine-toolbox-installation
# mamba update --name copernicusmarine copernicusmarine --yes
# copernicusmarine, version 1.2.2
cmd <- glue("
{cm} subset -i {dataset_id} \\
-x {b$xmin} -X {b$xmax} \\
-y {b$ymin} -Y {b$ymax} \\
-t {date} -T {date + months(1) - days(1)} \\
-z {depth - 0.01} -Z {depth + - 0.01} \\
-v {variable} \\
-o {dirname(nc)} -f {basename(nc)} \\
--username {user} --password '{pass}' \\
--force-download")
if (verbose)
print(cmd)
if (!file.exists(nc))
system(cmd, intern = TRUE)
r <- rast(nc)
# dimensions : 369, 353, 1 (nrow, ncol, nlyr)
# resolution : 0.08333334, 0.08333334 (x, y)
# extent : -135.2083, -105.7917, 18.45833, 49.20833 (xmin, xmax, ymin, ymax)
r_3857 <- leaflet::projectRasterForLeaflet(r, method = "bilinear")
# dimensions : 403, 313, 1 (nrow, ncol, nlyr)
# resolution : 10458.57, 10458.57 (x, y)
plet(r_3857, tiles = "Esri.WorldImagery")
```
## Interpolate larvae on Copernicus grid
### IDW(x,y)
```{r cm-larvae-idw}
# basename(nc) # cmems_mod_glo_phy_my_0.083deg_P1M-m__thetao__2022-04-01.nc"
r_cm <- rast(nc)
r_idw <- terra::interpIDW(
x = r_cm,
y = pts_ym |> terra::vect(),
"n_per_vol_log10",
# borrow from defaults in [calcofi4r:: pts_to_rast_idw()](https://github.com/CalCOFI/calcofi4r/blob/5e9508565a78785f8b374c3b8b5e2c4cdf559ed0/R/analyze.R#L236C5-L236C100)
radius = 1.5, power = 1.3, smooth = 0.2,
maxPoints = Inf, minPoints = 1, near = F, fill = NA) |>
mask(r_cm)
names(r_idw) <- "n_per_vol_log10_idw"
# time(r_idw) # "2005-04-01 UTC"
varnames(r_idw) <- ""
longnames(r_idw) <- ""
# units(r_idw) ""
# plot(r_idw)
pts_ymi <- r_idw |>
terra::extract(pts_ym, method = "simple", bind = T) |>
st_as_sf() |>
mutate(
dif_idw = n_per_vol_log10 - n_per_vol_log10_idw)
# pts_ymi$dif_idw |> summary()
# Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
# -2.24324 -0.28024 0.00000 0.01276 0.38674 1.76034 6
d_mi <- pts_ymi |>
st_drop_geometry() |>
filter(!is.na(n_per_vol_log10_idw))
rmse_idw <- with(
d_mi,
sqrt(
sum((n_per_vol_log10_idw - n_per_vol_log10)^2) /
nrow(d_mi) ) )
# [1] 0.7824212
mapView(
r_idw,
zcol = "n_per_vol_log10_idw",
col.regions = rev(RColorBrewer::brewer.pal(11,"Spectral"))) +
mapView(
pts_ymi,
zcol = "n_per_vol_log10",
col.regions = rev(RColorBrewer::brewer.pal(11,"Spectral"))) +
mapView(
pts_ymi,
zcol = "dif_idw",
col.regions = rev(RColorBrewer::brewer.pal(11,"Spectral")))
```
Root Mean Square Error: `r rmse_idw`
### GAM(x, y)
```{r cm-larvae-gam-xy}
librarian::shelf(mgcv)
# fit GAM
mdl_xy <- mgcv::gam(
n_per_vol_log10 ~ s(longitude, latitude, k = 60),
data = pts_ymi |> st_drop_geometry() |>
select(n_per_vol_log10, longitude, latitude),
method = "GCV.Cp", # method = "REML",
family = gaussian())
# summary(mdl_xy)
# Family: gaussian
# Link function: identity
#
# Formula:
# n_per_vol_log10 ~ s(longitude, latitude, k = 60)
#
# Parametric coefficients:
# Estimate Std. Error t value Pr(>|t|)
# (Intercept) 0.25360 0.06438 3.939 0.000117 ***
# ---
# Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#
# Approximate significance of smooth terms:
# edf Ref.df F p-value
# s(longitude,latitude) 12.91 18.26 11.79 <2e-16 ***
# ---
# Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#
# R-sq.(adj) = 0.526 Deviance explained = 55.7%
# GCV = 0.86605 Scale est. = 0.80397 n = 194
p_idw <- r_idw |>
as.points() |>
st_as_sf() |>
mutate(
longitude = sf::st_coordinates(geometry)[,1],
latitude = sf::st_coordinates(geometry)[,2])
# range(p_idw$n_per_vol_log10_idw)
# [1] -1.000000 2.413467
# predict with GAM
p_idw$n_per_vol_log10_xy <- predict.gam(
mdl_xy,
newdata = p_idw |>
st_drop_geometry() |>
select(longitude, latitude),
type = "response") # link") # "response")
# range(p_idw$n_per_vol_log10_xy) # -4.334638 0.183662
r_xy <- rasterize(
p_idw |>
st_drop_geometry() |>
select(longitude, latitude) |>
as.matrix(),
r_idw,
values = p_idw$n_per_vol_log10_xy)
# range(values(r_xy, na.rm=T)) # -4.334638 0.183662
# plot(r_xy)
names(r_xy) <- "n_per_vol_log10_xy"
pts_ymix <- r_xy |>
terra::extract(pts_ymi, method = "simple", bind = T) |>
st_as_sf() |>
mutate(
dif_xy = n_per_vol_log10 - n_per_vol_log10_xy)
# pts_ymix$dif_xy |> summary()
# Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
# -2.259856 -0.326382 -0.000841 0.024003 0.401710 2.446836 6
d_mix <- pts_ymix |>
st_drop_geometry() |>
filter(!is.na(n_per_vol_log10_idw))
rmse_xy <- with(
d_mix,
sqrt(
sum((n_per_vol_log10_xy - n_per_vol_log10)^2) /
nrow(d_mix) ) )
# [1] 0.8445961
mapView(
r_xy,
zcol = "n_per_vol_log10_xy",
col.regions = rev(RColorBrewer::brewer.pal(11,"Spectral"))) +
mapView(
pts_ymix,
zcol = "n_per_vol_log10",
col.regions = rev(RColorBrewer::brewer.pal(11,"Spectral"))) +
mapView(
pts_ymix,
zcol = "dif_xy",
col.regions = rev(RColorBrewer::brewer.pal(11,"Spectral")))
```
Root Mean Square Error: `r rmse_xy`
### GAM(x, y, temperature)
```{r cm-larvae-gam-xyt}
librarian::shelf(mgcv)
names(r_cm) <- "thetao"
pts_ymixt <- r_cm |>
terra::extract(pts_ymix, method = "simple", bind = T) |>
st_as_sf()
# fit GAM
mdl_xyt <- mgcv::gam(
n_per_vol_log10 ~ s(longitude, latitude, k = 60) + s(thetao),
data = pts_ymixt |> st_drop_geometry() |>
select(n_per_vol_log10, longitude, latitude, thetao),
method = "GCV.Cp", # method = "REML",
family = gaussian())
# summary(mdl_xyt)
# Family: gaussian
# Link function: identity
#
# Formula:
# n_per_vol_log10 ~ s(longitude, latitude, k = 60) + s(thetao)
#
# Parametric coefficients:
# Estimate Std. Error t value Pr(>|t|)
# (Intercept) 0.25181 0.06219 4.049 7.84e-05 ***
# ---
# Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#
# Approximate significance of smooth terms:
# edf Ref.df F p-value
# s(longitude,latitude) 12.767 18.199 12.505 <2e-16 ***
# s(thetao) 6.151 7.214 1.556 0.146
# ---
# Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#
# R-sq.(adj) = 0.576 Deviance explained = 61.9%
# GCV = 0.81327 Scale est. = 0.7271 n = 188
p_cm <- r_cm |>
mask(r_idw) |>
trim() |>
as.points() |>
st_as_sf() |>
mutate(
longitude = sf::st_coordinates(geometry)[,1],
latitude = sf::st_coordinates(geometry)[,2])
# range(p_idw$n_per_vol_log10_idw)
# [1] -1.000000 2.413467
# predict with GAM
p_cm$n_per_vol_log10_xyt <- predict.gam(
mdl_xyt,
newdata = p_cm |>
st_drop_geometry() |>
select(longitude, latitude, thetao),
type = "response") # link") # "response")
# range(p_cm$n_per_vol_log10_xyt) # -5.369945 1.165597
r_xyt <- rasterize(
p_cm |>
st_drop_geometry() |>
select(longitude, latitude) |>
as.matrix(),
r_idw,
values = p_cm$n_per_vol_log10_xyt)
# range(values(r_xyt, na.rm=T)) # -5.369945 1.165597
# plot(r_xy)
names(r_xyt) <- "n_per_vol_log10_xyt"
pts_ymixt <- r_xyt |>
terra::extract(pts_ymixt, method = "simple", bind = T) |>
st_as_sf() |>
mutate(
dif_xyt = n_per_vol_log10 - n_per_vol_log10_xyt)
# pts_ymixt$dif_xyt |> summary()
# Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
# -3.596383 -0.429587 -0.003567 0.002183 0.587119 2.562442 6
d_mixt <- pts_ymixt |>
st_drop_geometry() |>
filter(!is.na(n_per_vol_log10_idw))
rmse_xyt <- with(
d_mixt,
sqrt(
sum((n_per_vol_log10_xyt - n_per_vol_log10)^2) /
nrow(d_mixt) ) )
# rmse_xyt
# [1] 0.8054934
mapView(
r_xyt,
zcol = "n_per_vol_log10_xyt",
col.regions = rev(RColorBrewer::brewer.pal(11,"Spectral"))) +
mapView(
pts_ymixt,
zcol = "n_per_vol_log10",
col.regions = rev(RColorBrewer::brewer.pal(11,"Spectral"))) +
mapView(
pts_ymixt,
zcol = "dif_xyt",
col.regions = rev(RColorBrewer::brewer.pal(11,"Spectral")))
```
Root Mean Square Error: `r rmse_xyt`
